- populate units of COLONIES
- recreate the data table - clean up, add "S" to the first set A
- move assumptions out of all of the examples
- add row captions to highlight exceptions/assumptions
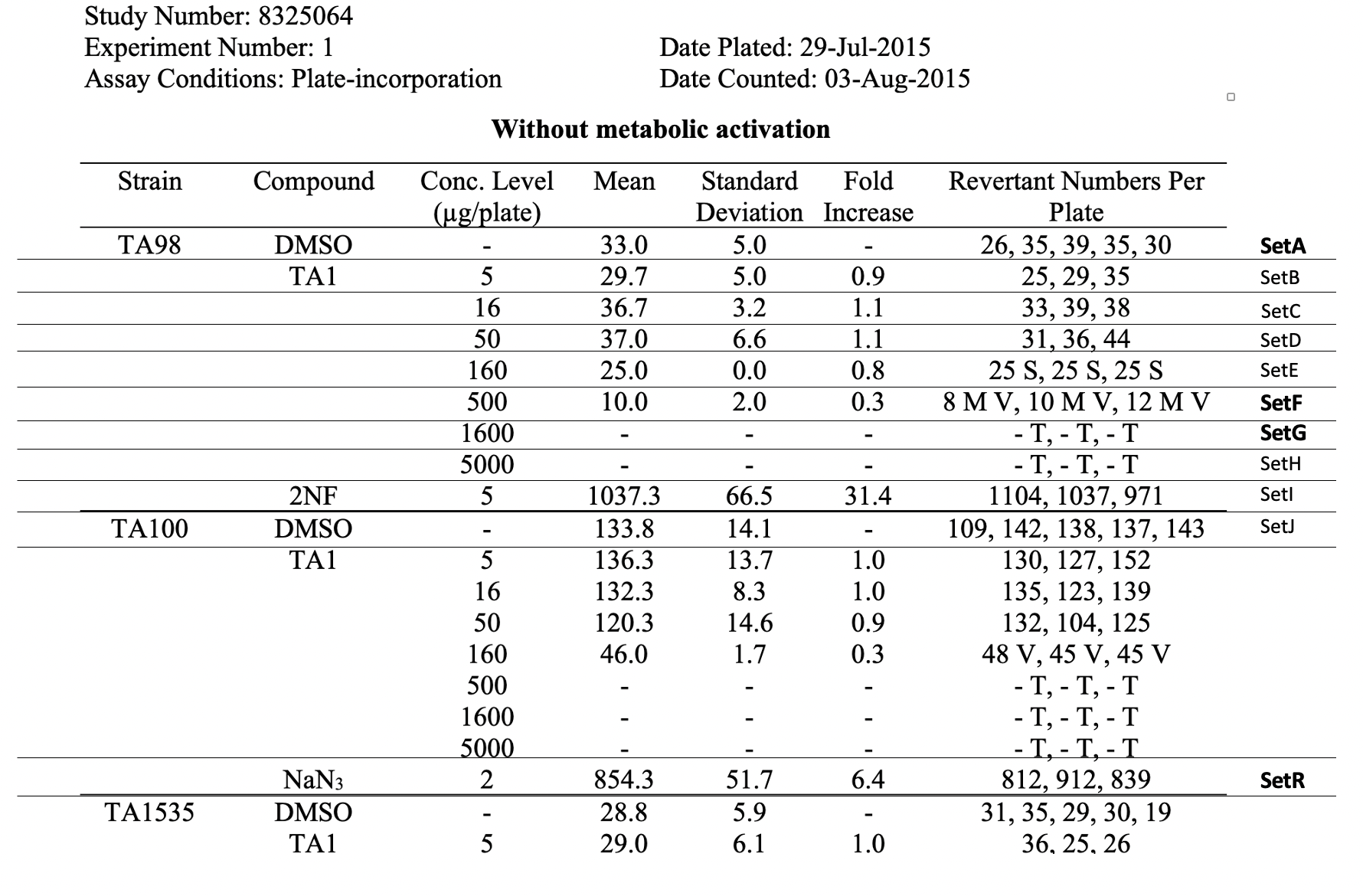
This is an example showing trial design and results from the in vitro bacterial reverse mutation test of Study #8325064. The bacterial reverse mutation test uses four different amino acid-requiring strains of Salmonella typhimurium (S. typhimurium) and one strain of Escherichia coli (E. coli) to detect point mutations, which involve substitution, addition or deletion of one or a few DNA base pairs. The principle of this bacterial reverse mutation test is that it detects chemicals that induce mutations which revert mutations present in the S typhimurium and E. coli tester strains and restore the functional capability of the bacteria to synthesize an essential amino acid. The revertant bacteria are detected by their ability to grow in the absence of the amino acid required by the parent tester strain.
Dataset Wrapper Debug Message
Row captions have been provided, but there doesn't seem to be a dataset to which the row captions would apply.
- How do we handle "toxic, no revertant colonies"? (LAK will add 6 records with GTSTAT = NOT DONE, GTREASND = "TOO MUCH CYTOTOXICITY,"
- When "-" in Mean, Std Dev, Fold Inc - refer to SEND team's marked-up table (GTSTAT = NOT DONE; variety of REASND)
| Rows 1-22: | Shows trial set parameters and trial set values that comprise the test conditions for the set SetA. SetA is the first row of the raw data table, see the row labeled "SetA" in the table, Raw Data for Study 8325064. |
|---|---|
| Rows 2-4: | Show two records for Alkaline Phosphatase that were collected 1 day apart and are expected to be reported together. Row 4 shows how to create a derived record (average of the Rows 2 and 3) and flag it as Derived (LBDRVFL = "Y") as well as the record to use as baseline (LBBLFL = "Y"). |
| Rows 6-7: | Show a suggested use of the LBSCAT variable. It could be used to further classify types of tests within a laboratory panel (e.g., "DIFFERENTIAL"). |
| Row 9: | Shows the proper use of the LBSTAT variable to indicate "NOT DONE," when the fact that the test was done was documented. |
| Row 12: | The subject had Cholesterol measured. The normal range for this test is <200 mg/dL. The value <200 may not be used in the normal range variables LBORNRHI or LBSTNRHI; however, a sponsor may decide, e.g., to enter "0" into LBORNRLO and "199" in LBORNRHI. The sponsor must define the appropriate value for all of the normal range variables. |
| Row 13: | Shows the use of LBUSCHFL to indicate that the test result was obtained from an unscheduled blood collection. In this case, the subject was moribund and a blood sample was taken prior to sacrifice. |
| Rows 1, 6: | Use records in the SUPPLB dataset example to show biological significance assigned by the investigator for test results. |
Row Captions Debug Message
Remember to keep number agreement between labels and captions.
Dataset Wrapper Debug Message
Please add a row column to your dataset.