Updates for SDTMIG 4.0
When this example is moved to the IG, make sure to update IS assumption #6: Measurements of cytokines, chemokines, and complement proteins should be represented in the Laboratory Test Results (LB) domain. Need to explain when to use IS vs LB for cytokine tests. See notes #3 below.
We recently received this request: https://loinc.org/95971-8/ --- SARS CoV-2 stimulated gamma interferon [Presence] in Blood. We also recieved requests for the following LOINC codes: 95974-2, 95972-6, and 95973-4. Upon further research on all 4 LOINC codes, they are referring to the IFNg Response Assay, which has been modeled in the LB domain in the TB-TAUG.
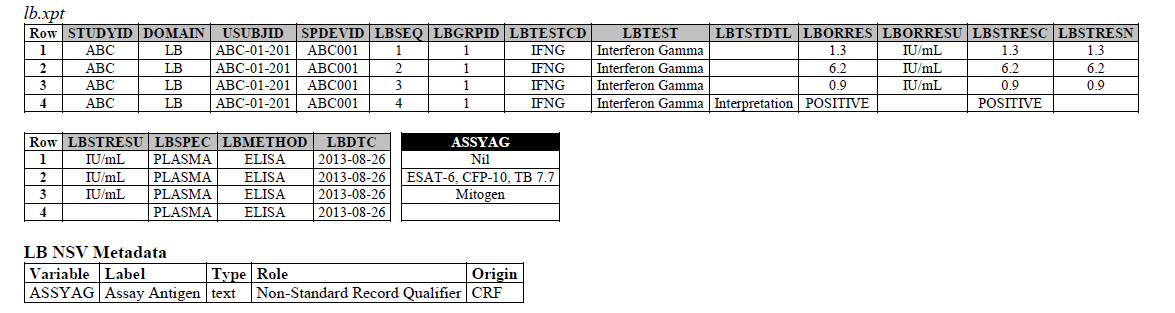
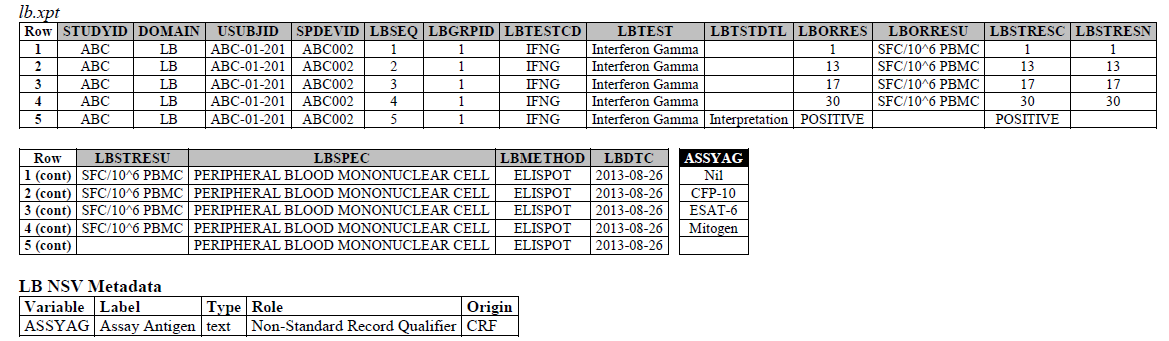
The IFNG Response test toward M. Tuberculosis is modeled in the LB domain, in the TB-TAUG as the following:
ELISA:
ELISPOT:
There are several concerns with this modeling:
- Where do you show that this challenge assay is done against M. Tuberculosis (or any microbe of interest, like SARS-CoV-2), where do you map M. Tb? - NHOID is NOT appropriate and should NOT be used here because in this case, the subject of study is NOT the Mycobacterium tuberculosis itself but whether someone has been previously exposed to M. Tuberculosis, by examining whether there is heightened Interferon-gamma activation. A positive result does not mean the person is currently infected with Mycobacterium tuberculosis, it may mean that the person had been previously infected by, or vaccinated against M. Tuberculosis. NHOID is used when you know the microorganism or a reference strain is present in the testing sample. M. Tuberculosis is mapped to ISCNDAGT to indicate that the stimulating agents are TB antigens.
When should NHOID be used?
NHOID, defined by the Non-host Organism Identifiers (OI) domain, should be used to map microorganisms that have been either experimentally determined in the course of a study or are previously known (e.g., lab strains used as reference in the study). In other words, NHOID is used when the study subject is the microorganism, and when the microorganism is present in the testing sample. In vaccine efficacy studies, a subject’s post-immunization sera is often incubated with a microbial strain of interest, where the functional capacities of the vaccine-induced antibodies are measured through whether the antibodies can effectively stop (from infection), neutralize, and kill the study microorganism of interest, in vitro. Examples of such tests include microneutralization, hemagglutination inhibition, and opsonophagocytic-killing assays. These are tests which measure the direct effect of the antimicrobial antibodies on the microorganism; therefore, said microorganism is the study subject and should be mapped to NHOID.
Tests that measure and quantify a subject's cellular and humoral immune responses to a microorganism or a vaccination agent—such as measurements of activated cytokine- or antibody–secreting cells, or cytokine response assays (e.g., interferon gamma response test)—are biological measurements about the human subject. Because these are not assessments about the microorganism itself, NHOID should not be used.
- CP domain has developed a new standard variable called Test Condition Agent/CNDAGT (CP Specification), which is used to represent stimulating agents, like the values in ASSYAG above, should this variable be added to LB, or IS? TEAM AGREES
- Lastly, Interferon-Gamma Response Assay is a classic immunological test, is it correct to map this to the LB domain?
- Can we model this in the IS domain, see example dataset below. Yes - TEAM AGREES
- If we model this test in IS, how do we distinguish it from the INFg test in lab. If we are running a routine lab test looking for IFNg levels it goes to LB. If we are running IFNg challenge/response test toward a specific, known microorganism then it goes to IS, you will also need to use the ISCNDAGT variable to house the challenge agent name. Yes-TEAM AGREES
If we were to model this in IS:
ELISA:
Similarly if we were to model Jozef's request in IS for https://loinc.org/95971-8/ --- SARS CoV-2 stimulated gamma interferon [Presence] in Blood