| Dataset wrap |
|---|
| Rowcaps |
|---|
| Rows 1-2: | Show 2 records for TSPARMCD = "GLPTYP", using TSSEQ to indicate multiple records, since both GLP types apply for this example study. | | Row 3: | Shows that this study was conducted as a GLP study. | | Rows 4-5: | Show the study start date and study title. | | Rows 6-7: | Show the version of SEND Implementation Guide and version of Controlled Terminology used in this study. | | Row 8: | Shows the applicant's organization. | | Row 9: | Shows that the applicant's study reference ID is not applicable. | | Rows 10-13: | Show that TSGRPID has been used to link records (name, location, country) related to the test facility (TSGRPID = 1). The study director is associated with the test facility. | | Rows 14-16: | Show that TSGRPID (TSGRPID=2) has been used to link the information on the testing guideline followed on this study (TSTGDNAM, TSTGDORG, TSTGDVER). | | Shows the study type for this study. | | Shows that this study includes a Mammalian Cell Micronucleus Assay. | | Rows 19-20: | Show that the species is human and the cell line is TK6 lymphoblastoid in this study. |
|
|
| Expand |
|---|
| title | ts.xpt (trial summary, study level parameters) |
|---|
|
- Assumption: A Trial (study) can have more than one assay type
- Assumption: ASSAYID value of ALL indicates that it applies to all assays in the study
- BLEO & CPA - are positive controls, should this be included/where is this captured? — in results for the record / row?
- Where is TK6 cell type? is this test system (see below)
- needs to be allowed to vary down to the well level / result level
- For the items highlighted in orange - what is the lowest level where this could vary? e.g. at the level of a well in a 96-well plate, or a slide vs. assay level vs. product level
- Smoke Regime needs to be allowed to vary down to the well level / result level
- run - also at result level
- port - (smoking product on the machine, you assign a run number or port-number for multiple replicates depending on linear vs. rotary machines; machine bias), needs to be allowed to vary down to the well level / result level)
- replicate - (e.g., multiple samples from one vial), could be a replicate per exposure (e.g., 3 plates, multiple analyses for exposure)
- sample ID - typically a unique number assigned (at CRO) or supplied by sponsor - result level
- Smoke fraction - result level (e.g.tpm particulate vs. gas gvp)
| Row | STUDYID | ASSAYID | DOMAIN | TSSEQ | TSGRPID | TSPARMCD | TSPARM | TSVAL | TSVALNF |
|---|
| 1 | 123 |
|
ALL | TS | 1 |
| GLPTYP | Good Laboratory Practice Type | FDA |
| | 2 | 123 |
|
ALL | TS | 2 |
| GLPTYP | Good Laboratory Practice Type | OECD |
| | 3 | 123 |
|
ALL | TS | 1 |
| STSTDTC | Study Start Date | 2022-05-25 |
|
4ALL | | TS | 1 |
| STITLE | Study Title | Determination of the in vitro genotoxicity potential |
|
of 10 tobacco products in both the in vitro Micronucleus Assay and the in vitro Neutral Red Uptakeusing the in vitro Neutral Red Uptake assay |
| | 6 | 123 |
|
5 | 123 | ALL | TS | 1 |
| SNDIGVER | SEND Implementation Guide Version | TOBACCO IMPLEMENTATION GUIDE VERSION 1.0 |
|
6ALL | | TS | 1 |
| SNDCTVER | SEND Controlled Terminology Version | SEND Terminology 2021-09-30 |
|
7ALL | SSPONSORSponsor Organization Sponsor 8ALLSPREFIDSponsor's | Study Reference ID |
| NOT APPLICABLE |
|
9ALL | TS | 1 | 1 | TSTFNAM | Test Facility Name | Example |
|
Tox 10ALL | TS | 1 | 1 | TSTFLOC | Test Facility Location | 10 Somewhere Street, Montgomery, AL 10000 |
|
11ALL | TS | 1 | 1 | TFCNTRY | Test Facility Country | USA |
|
...ALL | | TS | 1 | 1 | STDIR | Study Director | Dr. R. Smith |
| | 14 | 123 |
|
ALLGLPFL | GLP Flag | Y | 123 | | 2 | TSTGDNAM | Testing Guideline Name | GUIDELINE FOR THE TESTING OF CHEMICALS No. 487 |
| | 15 | 123 |
|
MNvitASTD | Assay Standard | | 2 | TSTGDORG | Testing Guideline Organization | OECD |
| | 16 |
|
OECD Test No. 487 MNvit | ASTDV | | 2 | TSTGDVER | Testing Guideline |
|
Assay Standard 20160729MNvit | TS | 1 |
| SSTYP | Study Type | GENOTOXICITY IN VITRO |
| | 18 | 123 |
|
MNvitSSSTYP | Study Sub Type | In Vitro Micronucleus |
| GNTXAID | Genetic Toxicology Assay Identifier | MNvit |
| | 19 | 123 |
|
123 | MNvitHomo SapiensMNvit?? | Test System? Lymphoblastoid Suspension Cells123 | NRU | TS | 1 | ASTD | Assay Standard | NIH Publication No. 07-4519 | 123 | NRU | TS | 1 | ASTDV | Assay Standard Version | 2006-11 | 123 | NRU | TS | 1 | SSTYP | Study Type | GENOTOXICITY IN VITRO | 123 | NRU | TS | 1 | SSSTYP | Study Sub Type | In Vitro Neutral Red Uptake | 123 | NRU | TS | 1 | SPECIES | Species | Salmonella enterica | 123 | NRU | TS | 1 | STRAIN | Strain/Substrain | Salmonella enterica enterica | 123 | NRU | TS | 1 | ?? | Test System?? | Normal Human Keratinocyte | 123 | NRU | TS | 1 | REGIME | Smoking Regime | Traditional combustible | 123 | NRU | TS | 1 | RUNPRT | run-port number?? (always same for whole assay?) | 1-1 | 123 | NRU | TS | 1 | SMPLID | Sample ID | 030001 | 123 | NRU | TS | 1 | SMKFRC | Smoke Fraction | A | 123 | NRU | TS | 1 | REPNUM | Replicate Number | 1 |
|---|
| Expand |
|---|
|
- During CT definition/reviews will decide appropriate TXPARM and TXVAL; Treatment duration may be controlled; For now, we just include good example values based on our experience
- Assumption: The Trial Sets (TX) domain provides the list of distinct sets of subjects having different experimental factors, treatment factors, inherent characteristics, or distinct sponsor designations as specified in the trial design.
| Row | STUDYID | ASSAYID | DOMAIN | SETCD | | TXSEQ | TXPARMCD | TXPARM | TXVAL |
|---|
1 | 123 | NRU | TX | 1-1-A(plate-col-row in the 96-well plate) | CSC-10(through SLS-200) | PLATENUM. (e.g., 1 or 2) | 1 | 1 | 2 | 123 | NRU | TX | 1-1-A | CSC-10 | WELLNUM (e.g., 1-A through 10-H) | Number representing the location of the well in the 96-well plate | 1-A | 3 | 123 | NRU | TX | 1-1-A | CSC-10 | INTRVN | name of the intervention article | Cigarette Smoke Condensate | 4 | 123 | NRU | TX | 1-1-A | CSC-10 | ITVTYPE(CT e.g, Product, positive control, negative control) | type of intervention article | PRODUCT | 5 | 123 | NRU | TX | 1-1-A | CSC-10 | ITVCONC | intervention concentration | 10 | 6 | 123 | NRU | TX | 1-1-A | CSC-10 | ITVCONCU | intervention unit | ug/ml | 7 | 123 | NRU | TX | 1-1-B | CSC-10 | PLATENUM | Plate number | 1 | 8 | 123 | NRU | TX | 1-1-B | CSC-10 | WELLNUM | Number representing the location of the well in the 96-well plate | 2-A | 9 | 123 | NRU | TX | 1-1-B | CSC-10 | INTRVN | name of the intervention article | Cigarette Smoke Condensate | 10 | 123 | NRU | TX | 1-1-B | CSC-10 | ITVTYPE | type of intervention article | PRODUCT | 11 | 123 | NRU | TX | 1-1-B | CSC-10 | ITVCONC | intervention concentration | 10 | 12 | 123 | NRU | TX | 1-1-B | CSC-10 | ITVCONCU | intervention unit | ug/ml | ... | ... | 123 | NRU | TX | 1-10-H | SLS-10 | PLATENUM | Plate number | 1 | 123 | NRU | TX | 1-10-H | SLS-10 | WELLNUM | Number representing the location of the well in the 96-well plate | 10-H | 123 | NRU | TX | 1-10-H | SLS-10 | INTRVN | name of the intervention article | sodium laurel sulfate | 123 | NRU | TX | 1-10-H | SLS-10 | ITVTYPE | type of intervention article | POSITIVE CONTROL | 123 | NRU | TX | 1-10-H | SLS-10 | ITVCONC | intervention concentration | 200 | 123 | NRU | TX | 1-10-H | SLS-10 | ITVCONCU | intervention unit | ug/ml | ... | ... | 123 | NRU | TX | 2-10-H | SLS-10 | PLATENUM | Plate number | 2 | 123 | NRU | TX | 2-10-H | SLS-10 | WELLNUM | Number representing the location of the well in the 96-well plate | 10-H | 123 | NRU | TX | 2-10-H | SLS-10 | INTRVN | name of the intervention article | sodium laurel sulfate | 123 | NRU | TX | 2-10-H | SLS-10 | ITVTYPE | type of intervention article | POSITIVE CONTROL | 123 | NRU | TX | 2-10-H | SLS-10 | ITVCONC | intervention concentration | 200 | 123 | NRU | TX | 2-10-H | SLS-10 | ITVCONCU | intervention unit | ug/ml | 123 | MNvit | TX | A1 | METACT | Metabolic Activation (should there be two parms? Presence, type)? | +S9 | 123 | MNvit | TX | A1 | TRTDRTRG | Treatment Duration target. (how do we show 3-6 hour range? start/end, target and tolerance?, one text field not-analyzable) | 3 | 123 | MNvit | TX | 123 | MNvit | TX | TRTDRTOL | Treatment Duration Tolerance | 123 | MNvit | TX | TRTDURU | Treatment Duration Unit (this is for both TRTDURT, TRTDURTOL) | H | 123 | MNvit | TX | INTRVN | name of the intervention article | Bleomycin or Cyclophosphamid A | 123 | MNvit | TX | ITVTYPE | type of intervention article | choices of values: product; negative control; positive control | 123 | MNvit | TX | A1 | ITVCONC | Concentration of i a | 0 | 123 | MNvit | TX | A1 | ITVCONCU | Concentration Unit | ug/ml | | Expand |
|---|
| title | gt.xpt (similar to LB) |
|---|
|
 Image Removed
Image Removed Image Removed
Image Removed
| Row | STUDYID | ASSAYID | DOMAIN | TXCD | GTSEQ | GTTESTCD | GTTEST | GTCELLEV
(cells evaluated) | GTORRES | GTORRESU | GTSTRESC | GTSTRESN | GTSTRESU | GTDTC | GTNOMDY | GTELTM | GTTPTREF |
|---|
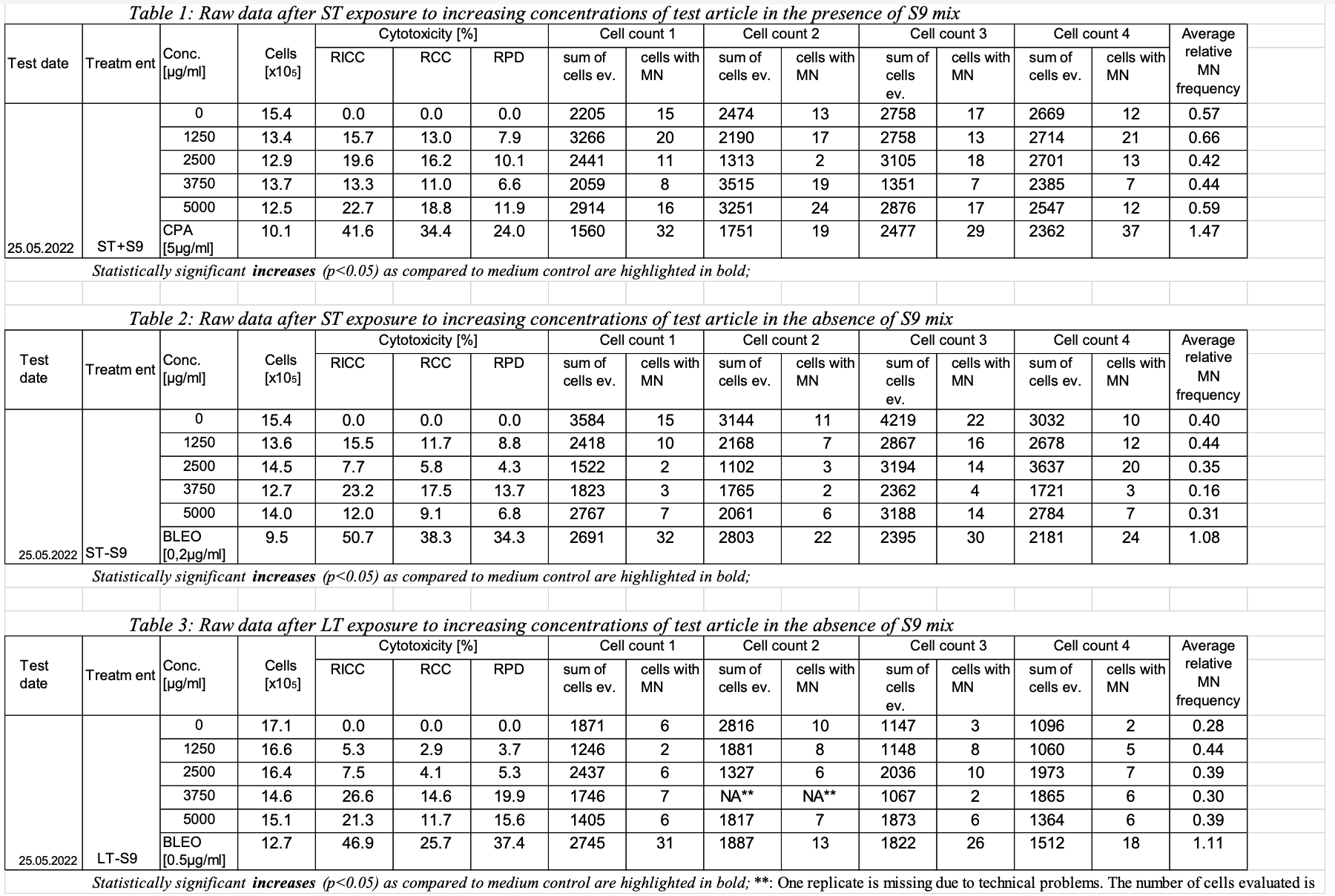
1 | 123 | NRU | GT | 1-1-A | 1 | RELABS | Relative Absorbance Reading | 100 | ug/ml | 100 | 100 | ug/ml | 2 | 123 | NRU | GT | 1-1-B | 1 | RELABS | Relative Absorbance Reading | 107 | ug/ml | 100 | 100 | ug/ml | 3 | 123 | NRU | GT | 1-1-C | 1 | RELABS | Relative Absorbance Reading | 98.6 | ug/ml | 100 | 100 | ug/ml | 54 | 123 | NRU | GT | 1-1-D | 1 | RELABS | Relative Absorbance Reading | 94.9 | ug/ml | 100 | 100 | ug/ml | 6 | 123 | NRU | GT | 1-1-E | 1 | RELABS | Relative Absorbance Reading | 111 | 7 | 123 | NRU | GT | 1-1-F | 1 | RELABS | Relative Absorbance Reading | 96.9 | 8 | 123 | NRU | GT | 1-1-G | 1 | RELABS | Relative Absorbance Reading | 105 | 9 | 123 | NRU | GT | 1-1-H | 1 | RELABS | Relative Absorbance Reading | 95.2 | 10 | 123 | NRU | GT | 1-2-A | 1 | RELABS | Relative Absorbance Reading | 83.0 | 11 | 123 | NRU | GT | 1-2-B | 1 | RELABS | Relative Absorbance Reading | 77.7 | ... | ... | 80 | 123 | NRU | GT | 1-10-H | 1 | RELABS | Relative Absorbance Reading | 0.791 | ... | ... | 160 | 123 | NRU | GT | 2-10-H | 1 | RELABS | Relative Absorbance Reading | 0.780 | 1 | 123 | MNvit | GT | A2 | 1 | RICC | Relative Increase in Cell Count | 134 | 15.7 | % | 15.7 | 15.7 | % | 2022-05-25 | 2 | 123 | MNvit | GT | A2 | 2 | RCC | Relative Cell Count | 134 | 13.0 | % | 13.0 | 13.0 | % | 2022-05-25 | 3 | 123 | MNvit | GT | A2 | 3 | RPD | Relative Population Doubling | 134 | 7.9 | % | 7.9 | 7.9 | % | 2022-05-25 | 4 | 123 | MNvit | GT | A2 | 4 | MNCELLS | Micronucleated Cells | 3266 | 20 | Cells | 20 | 20 | Cells | 2022-05-25 | 5 | 123 | MNvit | GT | A2 | 5 | MNCELLS | Micronucleated Cells | 2190 | 17 | Cells | 17 | 17 | Cells | 2022-05-25 | 6 | 123 | MNvit | GT | A2 | 6 | MNCELLS | Micronucleated Cells | 2758 | 13 | Cells | 13 | 13 | Cells | 2022-05-25 | 7 | 123 | MNvit | GT | A2 | 7 | MNCELLS | Micronucleated Cells | 2714 | 21 | Cells | 21 | 21 | Cells | 2022-05-25 | 8 | 123 | MNvit | GT | A2 | 8 | AVGREL | Average Relative MN Frequency | 0.66 | % | 0.66 | 0.66 | % | 2022-05-25 | 9 | 123 | MNvit | GT | A1 | 1 | RICC | Relative Increase in Cell Count | 154 | 0 | % | 0 | 0 | % | 2022-05-25 | 10 | 123 | MNvit | GT | A1 | 2 | RCC | Relative Cell Count | 154 | 0 | % | 0 | 0 | % | 2022-05-25 | 11 | 123 | MNvit | GT | A1 | 3 | RPD | Relative Population Doubling | 154 | 0 | % | 0 | 0 | % | 2022-05-25 | 12 | 123 | MNvit | GT | A1 | 4 | MNCELLS | Micronucleated Cells | 3266 | 15 | Cells | 15 | 15 | Cells | 2022-05-25 | 13 | 123 | MNvit | GT | A1 | 5 | MNCELLS | Micronucleated Cells | 2190 | 13 | Cells | 13 | 13 | Cells | 2022-05-25 | 14 | 123 | MNvit | GT | A1 | 6 | MNCELLS | Micronucleated Cells | 2758 | 17 | Cells | 17 | 17 | Cells | 2022-05-25 | 15 | 123 | MNvit | GT | A1 | 7 | MNCELLS | Micronucleated Cells | 2714 | 12 | Cells | 12 | 12 | Cells | 2022-05-25 | 16 | 123 | MNvit | GT | A1 | 8 | AVGREL | Average Relative MN Frequency | 0.57 | % | 0.57 | 0.57 | % | 2022-05-25 |
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
Rows 1-3, 8: | Show percentage result values that apply to GTREFID=C0. REFID=C0, as shown in the RELREF dataset, relates this data to the trial set in the first row of table 1 in the sample report table for study 123. | | Rows 4-7: | Show the 4 micronucleated cell counts for the observational units with GTREFID from C0-Count1 through C0-Count4, for which their relationship to test conditions (in tx.xpt) and experimental units (in relref.xpt) are shown in the RELREF dataset. | Rows 9-11, 16: | Show percentage result values that apply to GTREFID=C1250. REFID=C1250, as shown in the RELREF dataset, relates this data to the trial set in the second row of table 1 in the sample report table for study 123. | | Rows 12-15: | Show the 4 micronucleated cell counts for the observational units with GTREFID from C1250-Count1 through C1250-Count4, for which their relationship to test conditions (in tx.xpt) and experimental units (in relref.xpt) are shown in the RELREF dataset. |
|
| Dataset2 |
|---|
| Row | STUDYID | DOMAIN | GTSEQ | GTREFID | GTTESTCD | GTTEST | GTCELLEV | GTORRES | GTORRESU | GTCOLSRT | GTSTRESC | GTSTRESN | GTSTRESU | GTDTC |
|---|
| 1 | 123 | GT | 1 | C0 | | Relative Increase in Cell Count | 154 | 0 | % |
| 0 | 0 | % | 2022-05-25 | | 2 | 123 | GT | 2 | C0 | RCC | Relative Cell Count | 154 | 0 | % |
| 0 | 0 | % | 2022-05-25 | | 3 | 123 | GT | 3 | C0 | RPD | Relative Population Doubling | 154 | 0 | % |
| 0 | 0 | % | 2022-05-25 | | 4 | 123 | GT | 4 | C0-Count1 | MNCE | Micronucleated Cells | 2205 | 15 | |
| 15 | 15 |
| 2022-05-25 | | 5 | 123 | GT | 5 | C0-Count2 | MNCE | Micronucleated Cells | 2474 | 13 |
|
| 13 | 13 |
| 2022-05-25 | | 6 | 123 | GT | 6 | C0-Count3 | MNCE | Micronucleated Cells | 2758 | 17 |
|
| 17 | 17 |
| 2022-05-25 | | 7 | 123 | GT | 7 | C0-Count4 | MNCE | Micronucleated Cells | 2669 | 12 |
|
| 12 | 12 |
| 2022-05-25 | | 8 | 123 | GT | 8 | C0 | MNCECE | Micronucleated Cells/Total Cells |
| 0.57 | % |
| 0.57 | 0.57 | % | 2022-05-25 | | 9 | 123 | GT | 1 | C1250 | RICC | Relative Increase in Cell Count | 134 | 15.7 | % |
| 15.7 | 15.7 | % | 2022-05-25 | | 10 | 123 | GT | 2 | C1250 | RCC | Relative Cell Count | 134 | 13.0 | % |
| 13.0 | 13.0 | % | 2022-05-25 | | 11 | 123 | GT | 3 | C1250 | RPD | Relative Population Doubling | 134 | 7.9 | % |
| 7.9 | 7.9 | % | 2022-05-25 | | 12 | 123 | GT | 4 | C1250-Count1 | MNCE | Micronucleated Cells | 3266 | 20 |
|
| 20 | 20 |
| 2022-05-25 | | 13 | 123 | GT | 5 | C1250-Count2 | MNCE | Micronucleated Cells | 2190 | 17 |
|
| 17 | 17 |
| 2022-05-25 | | 14 | 123 | GT | 6 | C1250-Count3 | MNCE | Micronucleated Cells | 2758 | 13 |
|
| 13 | 13 |
| 2022-05-25 | | 15 | 123 | GT | 7 | C1250-Count4 | MNCE | Micronucleated Cells | 2714 | 21 |
|
| 21 | 21 |
| 2022-05-25 | | 16 | 123 | GT | 8 | C1250 | MNCECE | Micronucleated Cells/Total Cells |
| 0.66 | % |
| 0.66 | 0.66 | % | 2022-05-25 |
|
|
| Expand |
|---|
| title | te.xpt (trial elements) |
|---|
|
(Do we need this? a single element (treatment)?)
| Row | STUDYID | ASSAYID | DOMAIN | ETCD | ELEMENT |
|---|
| Expand |
|---|
|
(Do we need this? one arm for each trial set?)
| Row | STUDYID | ASSAYID | DOMAIN | ARMCD | ARM | TAETORD | ETCD | ELEMENT |
|---|
| Expand |
|---|
|
Not needed. | Row | STUDYID | DOMAIN | ENID (Entity ID) | RICC | RCC | RPD | Sum of cell ev. | Cells with MN | SMKFID (Smoke Fraction) | REPLCTID (Replicate Number) | PLATEID (Plate ID) | COLID (Column number) | ROWID (Row number) | SETCD (Set Code, TX) | RFSTDTC | RFENDTC | RFXSTDTC | RFXENDTC | RFCSTDTC | RFCENDTC | ARMCD
|---|