This is an example showing the report table, trial design, and results from a study's in vitro bacterial reverse mutation test using 4 different amino acid-requiring strains of S. typhimurium and 1 strain of E. coli to detect point mutations.
| Expand |
|---|
| title | Report Table for Example Study 8325064 |
|---|
|
This table shows the assay results for study 8325064, test article TA1, and 3 strains of salmonella (TA98, TA100, and TA1535) at varying concentrations. For brevity, the remaining tables (e.g., additional strains, samples prepared without metabolic activation) are not included. For ease of reference in the table below, each row has been labeled in the right-hand margin with the applicant-defined trial set label (e.g., SetA). For brevity, only the four bolded sets are represented in the following example datasets.
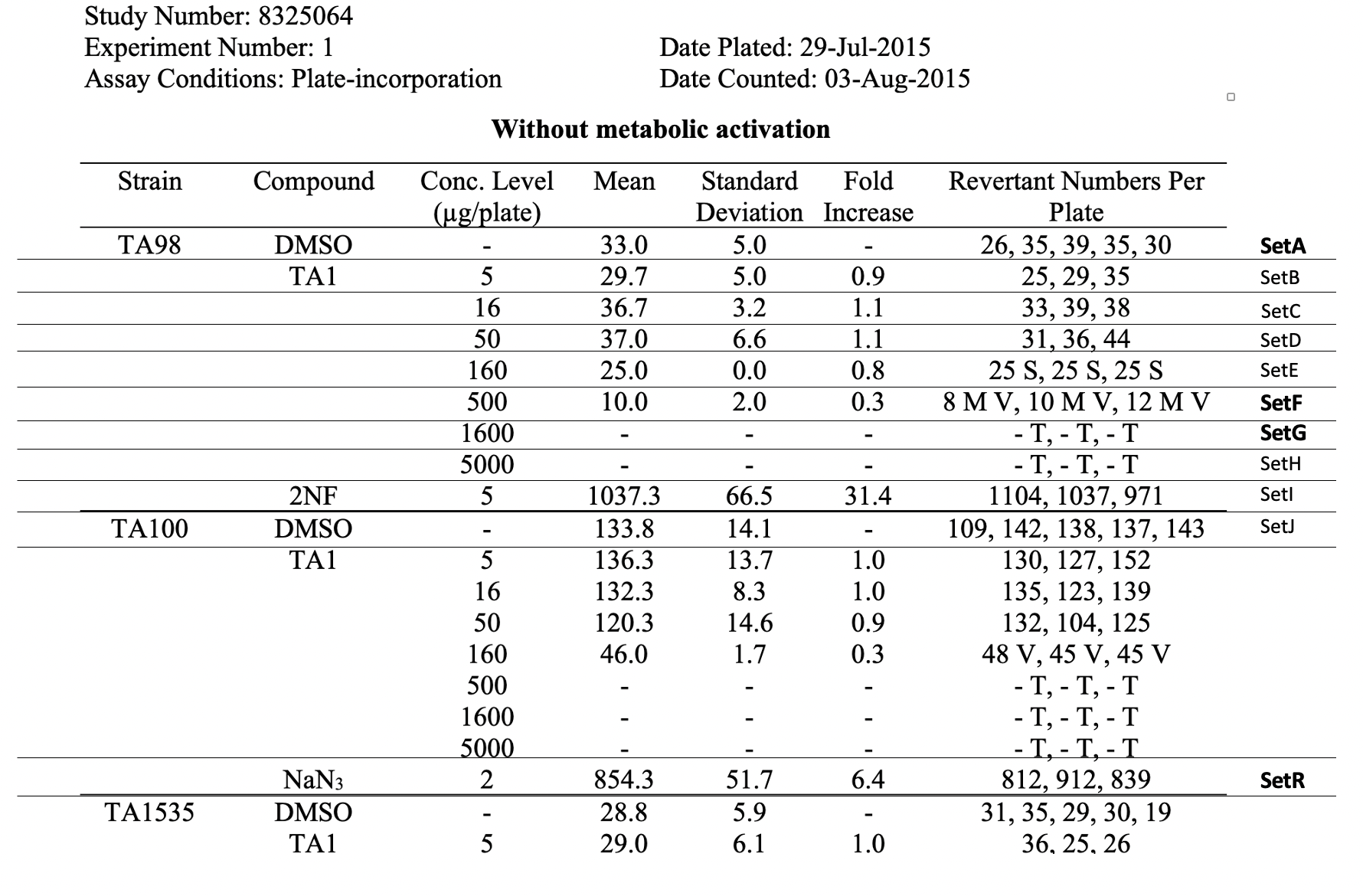 Image Added Image Added
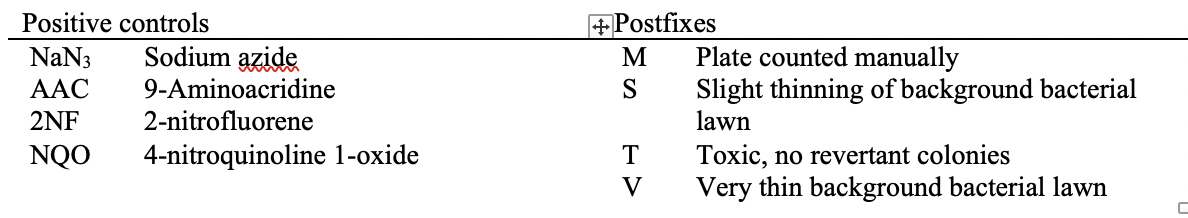 Image Added Image Added
|
This example Trial Summary (TS) dataset, shows many informational fields that provide context at the study level.
| Dataset wrap |
|---|
| Rowcaps |
|---|
| Rows 1-2: | Show 2 records for TSPARMCD = "GLPTYP", using TSSEQ to indicate multiple records, since both GLP types apply for this study. | | Row 3: | Shows that this study was conducted as a GLP study. | | Rows 4-5: | Show the study start date and study title. | | Rows 6-7: | Show the version of SEND Implementation Guide and version of Controlled Terminology used in this study. | | Row 8: | Shows the applicant's organization. | | Row 9: | Shows that the applicant's study reference ID is not applicable. | | Rows 10-13: | Show that TSGRPID has been used to link records (name, location, country) related to the test facility (TSGRPID=1). The study director is associated with the test facility. | | Rows 14-16: | Show that TSGRPID (TSGRPID=4) has been used to link the information on the testing guideline followed on this study (TSTGDNAM, TSTGDORG, TSTGDVER). | | Shows the study type for this study. | | Shows that this study includes a Bacterial Reverse Mutation Assay. | | Rows 19-27: | Show that TSGRPID (TSGRPID = 2) has been used to link the information for 1 species (salmonella) with the 4 different strains and cell lines that are tested in this study. | | Rows 28-30: | Show that TSGRPID (TSGRPID = 3) has been used to link the information for 1 species (E. coli) with the strain and cell line that is tested in this study. |
|
| Dataset2 |
|---|
| Row | STUDYID | DOMAIN | TSSEQ | TSGRPID | TSPARMCD | TSPARM | TSVAL | TSVALNF |
|---|
| 1 | 8325064 | TS | 1 |
| GLPTYP | Good Laboratory Practice Type | FDA |
| | 2 | 8325064 | TS | 2 |
| GLPTYP | Good Laboratory Practice Type | OECD |
| | 3 | 8325064 | TS | 1 |
| GLPFL | GLP Flag | N |
| | 4 | 8325064 | TS | 1 |
| STSTDTC | Study Start Date | 2015-07-29 |
| | 5 | 8325064 | TS | 1 |
| STITLE | Study Title | The Bacterial Reverse Mutation Test, Study 8325064-1 |
| | 6 | 8325064 | TS | 1 |
| SNDIGVER | SEND Implementation Guide Version | TOBACCO IMPLEMENTATION GUIDE VERSION 1.0 |
| | 7 | 8325064 | TS | 1 |
| SNDCTVER | SEND Controlled Terminology Version | SEND Terminology 2021-09-30 |
| | 8 | 8325064 | TS | 1 |
| APPLCNT | Applicant | Example Applicant Inc. |
| | 9 | 8325064 | TS | 1 |
| APREFID | Applicant's Study Reference ID |
| NOT APPLICABLE | | 10 | 8325064 | TS | 1 | 1 | TSTFNAM | Test Facility Name | Example Tox Lab Name |
| | 11 | 8325064 | TS | 1 | 1 | TSTFLOC | Test Facility Location | 10 Somewhere Street, Montgomery, AL 10000 |
| | 12 | 8325064 | TS | 1 | 1 | TFCNTRY | Test Facility Country | USA |
| | 13 | 8325064 | TS | 1 | 1 | STDIR | Study Director | Dr. R. Smith |
| | 14 | 8325064 | TS | 1 | 4 | TSTGDNAM | Testing Guideline Name | Test NO. 471 |
| | 15 | 8325064 | TS | 1 | 4 | TSTGDORG | Testing Guideline Organization | OECD |
| | 16 | 8325064 | TS | 1 | 4 | TSTGDVER | Testing Guideline Version | 2020-06-29 |
| | 17 | 8325064 | TS | 1 |
| SSTYP | Study Type | GENOTOXICITY IN VITRO |
| | 18 | 8325064 | TS | 1 |
| | Genetic Toxicology Assay Identifier | |
| | 19 | 8325064 | TS | 1 | 2 | | Species | |
| | 20 | 8325064 | TS | 1 | 2 | STRAIN | Strain/Substrain | TA98 |
| | 21 | 8325064 | TS | 2 | 2 | STRAIN | Strain/Substrain | TA100 |
| | 22 | 8325064 | TS | 3 | 2 | STRAIN | Strain/Substrain | TA1535 |
| | 23 | 8325064 | TS | 4 | 2 | STRAIN | Strain/Substrain | TA1537 |
| | 24 | 8325064 | TS | 1 | 2 | CELLLN | Cell Line | TA 98 hisD3052; rfa-; uvrB- |
| | 25 | 8325064 | TS | 2 | 2 | CELLLN | Cell Line | TA 100 hisG46; rfa-; uvrB- |
| | 26 | 8325064 | TS | 3 | 2 | CELLLN | Cell Line | TA 1535 hisG46; rfa-; uvrB- |
| | 27 | 8325064 | TS | 4 | 2 | CELLLN | Cell Line | TA 1537 hisC3076; rfa-; uvrB- |
| | 28 | 8325064 | TS | 2 | 3 | SPECIES | Species | |
| | 29 | 8325064 | TS | 5 | 3 | STRAIN | Strain/Substrain | WP2 uvrA pKM101 |
| | 30 | 8325064 | TS | 5 | 3 | CELLLN | Cell Line | trpE uvrA |
|
|
|
This example Trial Set (TX) dataset shows information about the test conditions for SetA and SetR in this study. For brevity, the dataset does not show information for SetF, SetG, or any other sets. A fully formed TX dataset for this example study would include information about the test conditions for all sets.
Note that there are three trial set parameters that link to other important datasets: SPTOBID, APDEVID, and SMKRGM.
- SPTOBID (Applicant-Defined Tobacco Product ID) is used to uniquely identify the tobacco product. The value of SPTOBID (e.g., CIG01a) matches the value for SPTOBID in all the TOPARMCD-TOVAL pairs in the Tobacco Product Identifiers and Descriptors (TO) dataset example in Section 3.1.2, Product Design Parameters and Conformance Testing, Example 1. The TOPARMCD-TOVAL pairs identify this unique product, CIG01a. The TO domain is described in Section 2.8.8.1, Tobacco Product Identifiers and Descriptors (TO).
- The value of APDEVID (e.g., PUFFMASTER3K) matches the value of SPDEVID in all the DIPARMCD-DIVAL pairs that identify this unique device in the DI dataset. SPDEVID is the applicant-defined device identifier that is used to uniquely identify the device in the Unique Device Identification (DI) dataset (see also Section 2.8.9.7, SEND Device Identifiers (DI)). This is shown in the DI dataset in Section 3.1.3.2, HPHCs, Other Constituents, and Smoking/Vaping Regimens, Example 1.
- SMKRGM serves as a link to the Device-In Use Properties (DU) domain (see also Section 2.8.9.8, SEND Device-In-Use (DU)), where a matching value of SMKRGM indicates parameters of the smoking regimen performed by the smoking machine, as in the DU dataset in Section 3.1.3.2, HPHCs, Other Constituents, and Smoking/Vaping Regimens, Example 2.
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
| Rows 1-22: | Show trial set parameters and trial set values that are the test conditions specific to set SetA which is the vehicle control (i.e., Strain of TA98, vehicle control with concentration value of 0). SetA is associated with the first row, labeled "SetA", in the report table for example study 8325064. | | Rows 23-44: | Show trial set parameters and trial set values that comprise the test conditions for the set SetF which is the strain of TA98 with a concentration value of 500 µg/plate. SetF is associated with the sixth row, labeled "SetF", in the report table for example study 8325064. | | Rows 45-66: | Show trial set parameters and trial set values that comprise the test conditions for the set SetG which is the strain of TA98 with a concentration value of 1600 µg/plate. SetG is associated with the seventh row, labeled "SetG", in the report table for example study 8325064. | | Rows 67-88: | Show trial set parameters and trial set values that comprise the test conditions for the set SetR which is the strain of TA100, positive control with a concentration value of 2 µg/plate. SetR is associated with the eighteenth row, labeled "SetR", in the report table for example study 8325064. |
|
|
- How do we handle "toxic, no revertant colonies"? (LAK will add 6 records with GTSTAT = NOT DONE, GTREASND = "TOO MUCH CYTOTOXICITY,"
- When "-" in Mean, Std Dev, Fold Inc - refer to SEND team's marked-up table (GTSTAT = NOT DONE; variety of REASND)
- populate units of COLONIES
- recreate the data table - clean up, add "S" to the first set A
- move assumptions out of all of the examples
- add row captions to highlight exceptions/assumptions
This is an example showing trial design and results from the in vitro bacterial reverse mutation test of Study #8325064. The bacterial reverse mutation test uses four different amino acid-requiring strains of Salmonella typhimurium (S. typhimurium) and one strain of Escherichia coli (E. coli) to detect point mutations, which involve substitution, addition or deletion of one or a few DNA base pairs. The principle of this bacterial reverse mutation test is that it detects chemicals that induce mutations which revert mutations present in the S typhimurium and E. coli tester strains and restore the functional capability of the bacteria to synthesize an essential amino acid. The revertant bacteria are detected by their ability to grow in the absence of the amino acid required by the parent tester strain.
 Image Removed
Image Removed
...
| title | ts.xpt (trial summary, study level parameters) |
|---|
- Assumption: When the experimental unit is derived from the species and strain, such as a cell line, then TESTSYS should be supplied.
...
Row
...
STUDYID
...
DOMAIN
...
TSSEQ
...
TSGRPID
...
TSPARMCD
...
TSPARM
...
TSVAL
...
TSVALNF
...
1
...
2
...
3
...
4
...
The Bacterial Reverse Mutation Test, Study 8325064-1
...
5
...
6
...
7
...
8
...
9
...
10
...
11
...
12
...
13
...
14
...
15
...
16
...
17
...
18
...
19
...
20
...
21
...
22
...
23
...
24
...
| Expand |
|---|
| title | Raw Data Table for Study 8325064 |
|---|
|
 Image Removed Image Removed
|
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
| Rows 1-22: | Shows trial set parameters and trial set values that comprise the test conditions for the set SetA. SetA is the first row of the raw data table, see the row labeled "SetA" in the table, Raw Data for Study 8325064. |
| Rows 2-4: | Show two records for Alkaline Phosphatase that were collected 1 day apart and are expected to be reported together. Row 4 shows how to create a derived record (average of the Rows 2 and 3) and flag it as Derived (LBDRVFL = "Y") as well as the record to use as baseline (LBBLFL = "Y"). |
| Rows 6-7: | Show a suggested use of the LBSCAT variable. It could be used to further classify types of tests within a laboratory panel (e.g., "DIFFERENTIAL"). |
| Row 13: | Shows the use of LBUSCHFL to indicate that the test result was obtained from an unscheduled blood collection. In this case, the subject was moribund and a blood sample was taken prior to sacrifice. |
| Rows 1, 6: | Use records in the SUPPLB dataset example to show biological significance assigned by the investigator for test results. |
| Expand |
|---|
|
|
| Row | STUDYID | ASSAYID | DOMAIN | SETCD | SET | TXSEQ | TXPARMCD | TXPARM | TXVAL |
|---|
| 1 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 1 | SPECIES | Species | Salmonella typhimurium |
| 2 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 2 | STRAIN | Strain/Substrain | TA98 |
3 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 3 | METACT | Metabolic Activation | | 4 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 4 | METACTFL | Y/N presence of metabolic activation | N |
| 5 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 5 | TRTDMIN | Treatment Duration Minimum | 71.5 |
| 6 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 6 | TRTDTRG | Treatment Duration Target | 72 |
| 7 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 7 | TRTDMAX | Treatment Duration Maximum | 72.5 |
| 8 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 8 | TRTDU | Treatment Duration Unit | HOURS |
| 9 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 9 | INCBTMP | Incubation Temperature | 37 |
| 10 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 10 | INCBTMPU | Incubation Temperature Unit | C |
| 11 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 11 | HUMID | Atmospheric Relative Humidity Percent | 50 |
| 12 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 12 | ATMCO2 | Atmospheric CO2 Percent | 5 |
| 13 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 13 | SPTOBID | Sponsor defined tobacco identifier | CIG01a |
| 14 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 14 | EXPTYP | Exposure Type (See TIG NC workstream minutes 30-Jan here: Nonclinical) | Submerged |
| 15 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 15 | SAMTYP | Sample Type | Total Particulate Matter in PBS |
| 16 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 16 | INTRVN | name of the intervention article | TA1 |
| 17 | 8325064 | Ames | TX | SetA | A-TA98-C0 | 17 | ITVTYPE | type of intervention article | Vehicle Control |
18 | 8325064 | Ames | | Dataset2 |
|---|
| Row | STUDYID | DOMAIN | SETCD | SET | TXSEQ | TXPARMCD | TXPARM | TXVAL |
|---|
| 1 | 8325064 |
|
18ITVCONC | Concentration of intervention article | 0 | | SPECIES | Species | SALMONELLA TYPHIMURIUM | | 2 |
|
19Ames | 19ITVCONCU | Concentration Unit | ug/plate | | IVTDMIN | In vitro Treatment Duration Minimum | 71.5 | | 3 | 8325064 |
|
20 | 8325064 | Ames20TRTV| IVTDTRG | In vitro Treatment |
|
VehicleDMSO21Ames | 21SPDEVID | Sponsor defined device identifier | PUFFMASTER3K | | IVTDMAX | In vitro Treatment Duration Maximum | 72.5 | | 5 | 8325064 |
|
22 | 8325064 | Ames22DUREFID | Smoke Regimen | Medium Intensity Regimen | | IVTDU | In vitro Treatment Duration Unit | HOURS | | 6 | 8325064 |
|
23 | 8325064 | AmesSetFFC5001SPECIESSpecies24AmesSetFFC5002 | STRAIN | Strain/Substrain | | 7 | INCBTMPU | Incubation Temperature Unit | C | | 8 |
|
25Ames | SetFFC5003 | METACT | Metabolic Activation | 26 | 8325064 | | ATMRHP | Atmospheric Relative Humidity Percent | 50 | | 9 | 8325064 |
|
AmesSetFFC5004METACTFL | Y/N presence of metabolic activation | N | | ATMCO2P | Atmospheric CO2 Percent | 5 | | 10 | 8325064 |
|
27 | 8325064 | AmesSetFFC500TRTDRTOLTreatment Duration Tolerance | 28 | 8325064 | Ames | TX | SetF | F-TA98-C500 | TRTDURU | Treatment Duration Unit (this is for both TRTDURT, TRTDURTOL) | 8325064 | Ames | TX | SetF | F-TA98-C500 | INTRVN | name of the intervention article (Tobacco ProdA, Bleomycin or Cyclophosphamid A) | TA1 | 8325064 | Ames | TX | SetF | F-TA98-C500 | ITVTYPE | type of intervention article choices of values: product; negative control; positive control | Product | | SPTOBID | Applicant-defined tobacco identifier | CIG01a | | 11 | 8325064 | TX | SetA | A-TA98-C0 | 11 | EXPTYP | | Submerged | | 12 | 8325064 | TX | SetA | A-TA98-C0 | 12 | SAMTYP | Sample Type | Total Particulate Matter in PBS | | 13 | 8325064 | TX | SetA | A-TA98-C0 | 13 | APDEVID | Applicant-defined device identifier | PUFFMASTER3K | | 14 | 8325064 | TX | SetA | A-TA98-C0 | 14 | SMKRGM | Smoking Regimen | NON-INTENSE REGIMEN | | 15 | 8325064 | TX | SetA | A-TA98-C0 | 15 |
|
8325064 | Ames | TX | SetF | F-TA98-C500 | ITVCONC | Concentration of intervention article | 500 | 8325064 | Ames | TX | SetF | F-TA98-C500 | ITVCONCU | Concentration Unit | ug/plate | 8325064 | Ames | TX | SetF | F-TA98-C500 | STRAIN | Strain/Substrain | TA98 | | 16 | 8325064 |
|
| TX | SetA | A-TA98-C0 | 16 | MTACTIND | Metabolic Activating Agent Name | NOT APPLICABLE | | 17 | 8325064 |
|
AmesSetFFC500REGIME | Smoking Regime | ISO Regime | ... | 8325064 | Ames | TX | SetG (row 7) do G not I!!! | METACT | Metabolic Activation (should there be two parms? Presence, type)? | None | 8325064 | Ames | TX | SetG | TRTDRTRG | Treatment Duration target. (how do we show 3-6 hour range? start/end, target and tolerance?, one text field not-analyzable) | 8325064 | Ames | TX | SetG | TRTDRTOL | Treatment Duration Tolerance | 8325064 | Ames | TX | SetG | TRTDURU | Treatment Duration Unit (this is for both TRTDURT, TRTDURTOL) | 8325064 | Ames | TX | SetG | INTRVN | name of the intervention article (Tobacco ProdA, Bleomycin or Cyclophosphamid A) | 2-Nitrofluorine | 8325064 | Ames | TX | SetG | ITVTYPE | type of intervention article choices of values: product; negative control; positive control | Positive Control | 8325064 | Ames | TX | SetG | ITVCONC | Concentration of intervention article | 5 | 8325064 | Ames | TX | SetG | ITVCONCU | Concentration Unit | ug/plate | 8325064 | Ames | TX | SetG | REGIME | Smoking Regime | ISO | ... | SetG | 8325064 | Ames | TX | SetR (Row 18) | METACT | Metabolic Activation (should there be two parms? Presence, type)? | None | 8325064 | Ames | TX | SetR | TRTDRTRG | Treatment Duration target. (how do we show 3-6 hour range? start/end, target and tolerance?, one text field not-analyzable) | 8325064 | Ames | TX | SetR | TRTDRTOL | Treatment Duration Tolerance | 8325064 | Ames | TX | SetR | TRTDURU | Treatment Duration Unit (this is for both TRTDURT, TRTDURTOL) | 8325064 | Ames | TX | SetR | INTRVN | name of the intervention article (Tobacco ProdA, Bleomycin or Cyclophosphamid A) | 4-Nitroquinoline-1-oxide | 8325064 | Ames | TX | SetR | ITVTYPE | type of intervention article choices of values: product; negative control; positive control | Positive Control | 8325064 | Ames | TX | SetR | ITVCONC | Concentration of intervention article | 2 | 8325064 | Ames | TX | SetR | ITVCONCU | Concentration Unit | ug/plate | 8325064 | Ames | TX | SetR | STRAIN | Strain/Substrain | TA98 | 8325064 | Ames | TX | SetR | REGIME | Smoking Regime | ISO | | Dataset wrap |
|---|
| Name | ou.xpt Observational and Experimental Unit Identifiers |
|---|
|
| Dataset wrap |
|---|
| Rowcaps |
|---|
NOTE: Observational Unit Identifiers (OBUIDs) are defined by the sponsor to uniquely identify the observational unit within an experimental unit. In this example OBUIDs are defined by the sponsor based on the test conditions (trial set) and the unit's location on one of multiple exposure/incubation plates.
| C0 | 17 | METACTFL | Presence of Metabolic Activation Flag | N | | 18 | 8325064 | TX | SetA | A-TA98-C0 | 18 | ITVNAM | Intervention Article Name | DMSO | | 19 | 8325064 | TX | SetA | A-TA98-C0 | 19 | ITVTYPE | Intervention Article Type | VEHICLE | | 20 | 8325064 | TX | SetA | A-TA98-C0 | 20 | ITVCONC | Intervention Article Concentration | 100 | | 21 | 8325064 | TX | SetA | A-TA98-C0 | 21 | ITVCONCU | Intervention Article Concentration Unit | % | | 22 | 8325064 | TX | SetA | A-TA98-C0 | 22 | TRTV | Treatment Vehicle | DMSO | | 23 | 8325064 | TX | SetF | F-TA98-C500 | 23 | SPECIES | Species | SALMONELLA TYPHIMURIUM | | 24 | 8325064 | TX | SetF | F-TA98-C500 | 24 | IVTDMIN | In vitro Treatment Duration Minimum | 71.5 | | 25 | 8325064 | TX | SetF | F-TA98-C500 | 25 | IVTDTRG | In vitro Treatment Duration Target | 72 | | 26 | 8325064 | TX | SetF | F-TA98-C500 | 26 | IVTDMAX | In vitro Treatment Duration Maximum | 72.5 | | 27 | 8325064 | TX | SetF | F-TA98-C500 | 27 | IVTDU | In vitro Treatment Duration Unit | HOURS | | 28 | 8325064 | TX | SetF | F-TA98-C500 | 28 | INCBTMP | Incubation Temperature | 37 | | 29 | 8325064 | TX | SetF | F-TA98-C500 | 29 | INCBTMPU | Incubation Temperature Unit | C | | 30 | 8325064 | TX | SetF | F-TA98-C500 | 30 | ATMRHP | Atmospheric Relative Humidity Percent | 50 | | 31 | 8325064 | TX | SetF | F-TA98-C500 | 31 | ATMCO2P | Atmospheric CO2 Percent | 5 | | 32 | 8325064 | TX | SetF | F-TA98-C500 | 32 | SPTOBID | Applicant-defined tobacco identifier | CIG01a | | 33 | 8325064 | TX | SetF | F-TA98-C500 | 33 | EXPTYP | | Submerged | | 34 | 8325064 | TX | SetF | F-TA98-C500 | 34 | SAMTYP | Sample Type | Total Particulate Matter in PBS | | 35 | 8325064 | TX | SetF | F-TA98-C500 | 35 | APDEVID | Applicant-defined device identifier | PUFFMASTER3K | | 36 | 8325064 | TX | SetF | F-TA98-C500 | 36 | SMKRGM | Smoking Regimen | NON-INTENSE REGIMEN | | 37 | 8325064 | TX | SetF | F-TA98-C500 | 37 | STRAIN | Strain/Substrain | TA98 | | 38 | 8325064 | TX | SetF | F-TA98-C500 | 38 | MTACTIND | Metabolic Activating Agent Name | NOT APPLICABLE | | 39 | 8325064 | TX | SetF | F-TA98-C500 | 39 | METACTFL | Presence of Metabolic Activation Flag | N | | 40 | 8325064 | TX | SetF | F-TA98-C500 | 40 | ITVNAM | Intervention Article Name | TA1 | | 41 | 8325064 | TX | SetF | F-TA98-C500 | 41 | ITVTYPE | Intervention Article Type | PRODUCT | | 42 | 8325064 | TX | SetF | F-TA98-C500 | 42 | ITVCONC | Intervention Article Concentration | 500 | | 43 | 8325064 | TX | SetF | F-TA98-C500 | 43 | ITVCONCU | Intervention Article Concentration Unit | ug/plate | | 44 | 8325064 | TX | SetF | F-TA98-C500 | 44 | TRTV | Treatment Vehicle | DMSO | | 45 | 8325064 | TX | SetG | G-TA98-C1600 | 45 | SPECIES | Species | SALMONELLA TYPHIMURIUM | | 46 | 8325064 | TX | SetG | G-TA98-C1600 | 46 | IVTDMIN | In vitro Treatment Duration Minimum | 71.5 | | 47 | 8325064 | TX | SetG | G-TA98-C1600 | 47 | IVTDTRG | In vitro Treatment Duration Target | 72 | | 48 | 8325064 | TX | SetG | G-TA98-C1600 | 48 | IVTDMAX | In vitro Treatment Duration Maximum | 72.5 | | 49 | 8325064 | TX | SetG | G-TA98-C1600 | 49 | IVTDU | In vitro Treatment Duration Unit | HOURS | | 50 | 8325064 | TX | SetG | G-TA98-C1600 | 50 | INCBTMP | Incubation Temperature | 37 | | 51 | 8325064 | TX | SetG | G-TA98-C1600 | 51 | INCBTMPU | Incubation Temperature Unit | C | | 52 | 8325064 | TX | SetG | G-TA98-C1600 | 52 | ATMRHP | Atmospheric Relative Humidity Percent | 50 | | 53 | 8325064 | TX | SetG | G-TA98-C1600 | 53 | ATMCO2P | Atmospheric CO2 Percent | 5 | | 54 | 8325064 | TX | SetG | G-TA98-C1600 | 54 | SPTOBID | Applicant-defined tobacco identifier | CIG01a | | 55 | 8325064 | TX | SetG | G-TA98-C1600 | 55 | EXPTYP | | Submerged | | 56 | 8325064 | TX | SetG | G-TA98-C1600 | 56 | SAMTYP | Sample Type | Total Particulate Matter in PBS | | 57 | 8325064 | TX | SetG | G-TA98-C1600 | 57 | APDEVID | Applicant-defined device identifier | PUFFMASTER3K | | 58 | 8325064 | TX | SetG | G-TA98-C1600 | 58 | SMKRGM | Smoking Regimen | NON-INTENSE REGIMEN | | 59 | 8325064 | TX | SetG | G-TA98-C1600 | 59 | STRAIN | Strain/Substrain | TA98 | | 60 | 8325064 | TX | SetG | G-TA98-C1600 | 60 | MTACTIND | Metabolic Activating Agent Name | NOT APPLICABLE | | 61 | 8325064 | TX | SetG | G-TA98-C1600 | 61 | METACTFL | Presence of Metabolic Activation Flag | N | | 62 | 8325064 | TX | SetG | G-TA98-C1600 | 62 | ITVNAM | Intervention Article Name | TA1 | | 63 | 8325064 | TX | SetG | G-TA98-C1600 | 63 | ITVTYPE | Intervention Article Type | Product | | 64 | 8325064 | TX | SetG | G-TA98-C1600 | 64 | ITVCONC | Intervention Article Concentration | 1600 | | 65 | 8325064 | TX | SetG | G-TA98-C1600 | 65 | ITVCONCU | Intervention Article Concentration Unit | ug/plate | | 66 | 8325064 | TX | SetG | G-TA98-C1600 | 66 | TRTV | Treatment Vehicle | DMSO | | 67 | 8325064 | TX | SetR | R-TA100-C2 | 67 | SPECIES | Species | SALMONELLA TYPHIMURIUM | | 68 | 8325064 | TX | SetR | R-TA100-C2 | 68 | IVTDMIN | In vitro Treatment Duration Minimum | 71.5 | | 69 | 8325064 | TX | SetR | R-TA100-C2 | 69 | IVTDTRG | In vitro Treatment Duration Target | 72 | | 70 | 8325064 | TX | SetR | R-TA100-C2 | 70 | IVTDMAX | In vitro Treatment Duration Maximum | 72.5 | | 71 | 8325064 | TX | SetR | R-TA100-C2 | 71 | IVTDU | In vitro Treatment Duration Unit | HOURS | | 72 | 8325064 | TX | SetR | R-TA100-C2 | 72 | INCBTMP | Incubation Temperature | 37 | | 73 | 8325064 | TX | SetR | R-TA100-C2 | 73 | INCBTMPU | Incubation Temperature Unit | C | | 74 | 8325064 | TX | SetR | R-TA100-C2 | 74 | ATMRHP | Atmospheric Relative Humidity Percent | 50 | | 75 | 8325064 | TX | SetR | R-TA100-C2 | 75 | ATMCO2P | Atmospheric CO2 Percent | 5 | | 76 | 8325064 | TX | SetR | R-TA100-C2 | 76 | SPTOBID | Applicant-defined tobacco identifier | CIG01a | | 77 | 8325064 | TX | SetR | R-TA100-C2 | 77 | EXPTYP | | Submerged | | 78 | 8325064 | TX | SetR | R-TA100-C2 | 78 | SAMTYP | Sample Type | Total Particulate Matter in PBS | | 79 | 8325064 | TX | SetR | R-TA100-C2 | 79 | APDEVID | Applicant-defined device identifier | PUFFMASTER3K | | 80 | 8325064 | TX | SetR | R-TA100-C2 | 80 | SMKRGM | Smoking Regimen | NON-INTENSE REGIMEN | | 81 | 8325064 | TX | SetR | R-TA100-C2 | 81 | STRAIN | Strain/Substrain | TA100 | | 82 | 8325064 | TX | SetR | R-TA100-C2 | 82 | MTACTIND | Metabolic Activating Agent Name | NOT APPLICABLE | | 83 | 8325064 | TX | SetR | R-TA100-C2 | 83 | METACTFL | Presence of Metabolic Activation Flag | N | | 84 | 8325064 | TX | SetR | R-TA100-C2 | 84 | ITVNAM | Intervention Article Name | NaN3 | | 85 | 8325064 | TX | SetR | R-TA100-C2 | 85 | ITVTYPE | Intervention Article Type | Positive Control | | 86 | 8325064 | TX | SetR | R-TA100-C2 | 86 | ITVCONC | Intervention Article Concentration | 2 | | 87 | 8325064 | TX | SetR | R-TA100-C2 | 87 | ITVCONCU | Intervention Article Concentration Unit | ug/plate | | 88 | 8325064 | TX | SetR | R-TA100-C2 | 88 | TRTV | Treatment Vehicle | DMSO |
|
|
REFID values are defined by the applicant to uniquely identify the observational unit within an experimental unit. In the simplified diagram of an assay procedure (below), the test tube that contains the possible mutagen is an example of an entity that would have a REFID indicating information at the level of trial set. The petri plate created from that test tube and used to count revertant numbers is an example of an entity that would be at an observational level.
This picture does not depict the more complicated procedures, trial sets, or observational units for the example study. For study 8325064, the applicant chose to define the REFIDs at the trial set level with a single character (e.g., A, F, G, R) and chose to define REFIDs at the observational level based on the location of each tube in 1 of multiple exposure/incubation plates (e.g, 6_1 for plate 6, position 1).
 Image Added
Image Added
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
| Row 1: | Shows the value of REFID=A. This REFID refers to the trial set with a SETCD of "SetA", as defined in the TX dataset. LEVEL=1 and LVLDESC="EXPERIMENTAL UNIT/TRIAL SET". TRIAL SET indicates this identifier relates to the entire trial set. EXPERIMENTAL UNIT indicates this identifier received the intervention article, DMSO. | | Rows 2-6: | Show the values of 5 observational units (1_1 through 1_5) that are within the parent experimental unit, REFID=A. | | Row 7: | Shows the value of REFID=F. This REFID refers to the trial set with a SETCD of "SetF", as defined in the TX dataset. LEVEL=1 and LVLDESC="EXPERIMENTAL UNIT/TRIAL SET". TRIAL SET indicates this identifier relates to the entire trial set. EXPERIMENTAL UNIT indicates this identifier received the intervention article, TA1. | | Rows 8-10: | Show the values of 3 observational units (6_1 through 6_3) that are within the parent experimental unit, REFID=F. | | Row 11: | Shows the value of REFID=G. This REFID refers to the trial set with a SETCD of "SetG", as defined in the TX dataset. LEVEL=1 and LVLDESC="EXPERIMENTAL UNIT/TRIAL SET". TRIAL SET indicates this identifier relates to the entire trial set. EXPERIMENTAL UNIT indicates this identifier received the intervention article, TA1. | | Rows 12-14: | Show the values of 3 observational units (7_1 through 7_3) that are within the parent experimental unit, REFID=G. | | Row 15: | Shows the value of REFID=R. This REFID refers to the trial set with a SETCD of "SetG", as defined in the TX dataset. LEVEL=1 and LVLDESC="EXPERIMENTAL UNIT/TRIAL SET". TRIAL SET indicates this identifier relates to the entire trial set. EXPERIMENTAL UNIT indicates this identifier received the intervention article, NaN3. | | Rows 16-18: | Show the values of 3 observational units (18_1 through 18_3) that are within the parent experimental unit, REFID=R. |
|
| Dataset2 |
|---|
| Row | STUDYID | SETCD | REFID | PARENT | LEVEL | LVLDESC |
|---|
1 | 8325064 | SetA | A |
| 1 | EXPERIMENTAL UNIT/TRIAL SET |
|---|
2 | 8325064 | SetA | 1_1 | A | 2 | OBSERVATIONAL UNIT |
|---|
3 | 8325064 | SetA | 1_2 | A | 2 | OBSERVATIONAL UNIT |
|---|
4 | 8325064 | SetA | 1_3 | A | 2 | OBSERVATIONAL UNIT |
|---|
5 | 8325064 | SetA | 1_4 | A | 2 | OBSERVATIONAL UNIT |
|---|
6 | 8325064 | SetA | 1_5 | A | 2 | OBSERVATIONAL UNIT |
|---|
7 | 8325064 | SetF | F |
| 1 | EXPERIMENTAL UNIT/TRIAL SET |
|---|
8 | 8325064 | SetF | 6_1 | F | 2 | OBSERVATIONAL UNIT |
|---|
9 | 8325064 | SetF | 6_2 | F | 2 | OBSERVATIONAL UNIT |
|---|
10 | 8325064 | SetF | 6_3 | F | 2 | OBSERVATIONAL UNIT |
|---|
11 | 8325064 | SetG | G |
| 1 | EXPERIMENTAL UNIT/TRIAL SET |
|---|
12 | 8325064 | SetG | 7_1 | G | 2 | OBSERVATIONAL UNIT |
|---|
13 | 8325064 | SetG | 7_2 | G | 2 | OBSERVATIONAL UNIT |
|---|
| 14 | 8325064 | SetG | 7_3 | G | 2 | OBSERVATIONAL UNIT | | 15 | 8325064 | SetR | R |
| 1 | EXPERIMENTAL UNIT/TRIAL SET | | 16 | 8325064 | SetR | 18_1 | R | 2 | OBSERVATIONAL UNIT | | 17 | 8325064 | SetR | 18_2 | R | 2 | OBSERVATIONAL UNIT | | 18 | 8325064 | SetR | 18_3 | R | 2 | OBSERVATIONAL UNIT |
|
|
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
| Rows 1-5: | Show the number of revertant colonies per plate collected for each of 5 observational units, GTREFID=1_1 through 1_5 (see description in the RELREF dataset). | | Rows 6, 7: | Show summary values collected (mean, standard deviation) for GTREFID=A that apply to the entire trial set, SetA, as indicated by LEVEL=1 and LVLDESC=TRIAL SET for this REFID as shown in the RELREF dataset. | | Rows 8-13: | Revertent colonies were counted for each of 3 plates/observational units (GTREFID=6_1 through 6_3) and each value is associated with a record to show a postfix code of "V" (Very thin background bacterial lawn). | | Rows 14-16: | Show summary values collected (mean, standard deviation, fold increase) for GTREFID=F that apply to the entire trial set, SetF, as indicated by LEVEL=1 and LVLDESC=TRIAL SET for this REFID in the RELREF dataset. | | Rows 17-19: | Show 3 plates/observational units (GTREFID=7_1 through 7_3) where no revertant colonies were counted due to too much cytotoxicity and a postfix code of "T". | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| columnIds | issuekey,summary,issuetype,created,updated,duedate,assignee,reporter,priority,status,resolution |
|---|
| columns | key,summary,type,created,updated,due,assignee,reporter,priority,status,resolution |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-668 |
|---|
|
| | Rows 20-22: | Show the number of revertant colonies per plate collected for each of 3 observational units (GTREFID=18_1 through 18_3). | | Rows 23-25: | Show summary values collected (mean, standard deviation, fold increase) for GTREFID=R that apply to the entire trial set, SetR, as indicated by LEVEL=1 and LVLDESC=TRIAL SET for this REFID in the RELREF dataset. |
|
| Dataset2 |
|---|
| Row | STUDYID | DOMAIN | GTSEQ | GTREFID | GTTESTCD | GTTEST | GTORRES | GTORRESU | GTCOLSRT | GTSTRESC | GTSTRESN | GTSTRESU | GTSTAT | GTREASND | GTMETHOD | GTDTC |
|---|
| 1 | 8325064 | GT | 1 | 1_1 | RPP | Revertant Colony Numbers Per Plate | 26 |
|
| 26 | 26 | |
|
| INSTRUMENT COUNTED | 2015-08-03 | | 2 | 8325064 | GT | 2 | 1_2 | RPP | Revertant Colony Numbers Per Plate | 35 |
|
| 35 | 35 |
|
|
| INSTRUMENT COUNTED | 2015-08-03 | | 3 | 8325064 | GT | 3 | 1_3 | RPP | Revertant Colony Numbers Per Plate | 39 |
|
| 39 | 39 |
|
|
| INSTRUMENT COUNTED | 2015-08-03 | | 4 | 8325064 | GT | 4 | 1_4 | RPP | Revertant Colony Numbers Per Plate | 35 |
|
| 35 | 35 |
|
|
| INSTRUMENT COUNTED | 2015-08-03 | | 5 | 8325064 | GT | 5 | 1_5 | RPP | Revertant Colony Numbers Per Plate | 30 |
|
| 30 | 30 |
|
|
| INSTRUMENT COUNTED | 2015-08-03 | | 6 | 8325064 | GT | 6 | A | RPP | Revertant Colony Numbers Per Plate | 33.0 |
| MEAN | 33.0 | 33.0 |
|
|
|
| 2015-08-03 | | 7 | 8325064 | GT | 7 | A | RPP | Revertant Colony Numbers Per Plate | 5.0 |
| STANDARD DEVIATION | 5.0 | 5.0 |
|
|
|
| 2015-08-03 | | 8 | 8325064 | GT | 8 | 6_1 | RPP | Revertant Colony Numbers Per Plate | 8 |
|
| 8 | 8 |
|
|
| MANUALLY COUNTED | 2015-08-03 | | 9 | 8325064 | GT | 9 | 6_1 | CYTOTOX | Cytotoxicity | Very thin background bacterial lawn |
|
| V |
|
|
|
|
| 2015-08-03 | | 10 | 8325064 | GT | 10 | 6_2 | RPP | Revertant Colony Numbers Per Plate | 10 |
|
| 10 | 10 |
|
|
| MANUALLY COUNTED | 2015-08-03 | | 11 | 8325064 | GT | 11 | 6_2 | CYTOTOX | Cytotoxicity | Very thin background bacterial lawn |
|
| V |
|
|
|
|
| 2015-08-03 | | 12 | 8325064 | GT | 12 | 6_3 | RPP | Revertant Colony Numbers Per Plate | 12 |
|
| 12 | 12 |
|
|
| MANUALLY COUNTED | 2015-08-03 | | 13 | 8325064 | GT | 13 | 6_3 | CYTOTOX | Cytotoxicity | Very thin background bacterial lawn |
|
| V |
|
|
|
|
| 2015-08-03 | | 14 | 8325064 | GT | 14 | F | RPPMEAN | Mean Rev Colony Num Per Plate | 10.0 |
| MEAN | 10.0 | 10.0 |
|
|
|
| 2015-08-03 | | 15 | 8325064 | GT | 15 | F | RPPSTDDV | Std Dev Rev Colony Num Per Plate | 2.0 |
| STANDARD DEVIATION | 2.0 | 2.0 |
|
|
|
| 2015-08-03 | | 16 | 8325064 | GT | 16 | F | RPPFLDIC | Fold Increase Rev Colony Num Per Plate | 0.3 |
| FOLD INCREASE | 0.3 | 0.3 |
|
|
|
| 2015-08-03 | | 17 | 8325064 | GT | 17 | 7_1 | CYTOTOX | Cytotoxicity | Toxic No Revertant Colonies |
|
| T |
|
| NOT DONE | TOO MUCH CYTOTOXICITY |
| 2015-08-03 | | 18 | 8325064 | GT | 18 | 7_2 | CYTOTOX | Cytotoxicity | Toxic No Revertant Colonies |
|
| T |
|
| NOT DONE | TOO MUCH CYTOTOXICITY |
| 2015-08-03 | | 19 | 8325064 | GT | 19 | 7_3 | CYTOTOX | Cytotoxicity | Toxic No Revertant Colonies |
|
| T |
|
| NOT DONE | TOO MUCH CYTOTOXICITY |
| 2015-08-03 | | 20 | 8325064 | GT | 20 | 18_1 | RPP | Revertant Colony Numbers Per Plate | 812 |
|
| 812 | 812 |
|
|
| INSTRUMENT COUNTED | 2015-08-03 | | 21 | 8325064 | GT | 21 | 18_2 | RPP | Revertant Colony Numbers Per Plate | 912 |
|
| 912 | 912 |
|
|
| INSTRUMENT COUNTED | 2015-08-03 | | 22 | 8325064 | GT | 22 | 18_3 | RPP | Revertant Colony Numbers Per Plate | 839 |
|
| 839 | 839 |
|
|
| INSTRUMENT COUNTED | 2015-08-03 | | 23 | 8325064 | GT | 23 | R | RPPMEAN | Mean Rev Colony Num Per Plate | 854.3 |
| MEAN | 854.3 | 854.3 |
|
|
|
| 2015-08-03 | | 24 | 8325064 | GT | 24 | R | RPPSTDDV | Std Dev Rev Colony Num Per Plate | 51.7 |
| STANDARD DEVIATION | 51.7 | 51.7 |
|
|
|
| 2015-08-03 | | 25 | 8325064 | GT | 25 | R | RPPFLDIC | Fold Increase Rev Colony Num Per Plate | 6.4 |
| FOLD INCREASE | 6.4 | 6.4 |
|
|
|
| 2015-08-03 |
|
|
| Rows 1-5: | Show the values for OBUID within the experimental unit identifier for Set A, as defined in the trial set dataset. |
| Rows 6-8: | Show the values for OBUID within the experimental unit identifier for Set F, as defined in the trial set dataset. |
| Rows 9-11: | Show the values for OBUID within the experimental unit identifier for Set G, as defined in the trial set dataset. |
| Row 12-14: | Show the values for OBUID within the experimental unit identifier for Set R, as defined in the trial set dataset. |
| Dataset2 |
|---|
| Row | STUDYID | ASSAYID | DOMAIN | SETCD | EUID | OBUID |
|---|
| 1 | 8325064 | Ames | OU | SetA | A | 0_1 | | 2 | 8325064 | Ames | OU | SetA | A | 0_2 | | 3 | 8325064 | Ames | OU | SetA | A | 0_3 | | 4 | 8325064 | Ames | OU | SetA | A | 0_4 | | 5 | 8325064 | Ames | OU | SetA | A | 0_5 | 6 | 8325064 | Ames | OU | SetF | F | 6_1 | | 7 | 8325064 | Ames | OU | SetF | F | 6_2 | | 8 | 8325064 | Ames | OU | SetF | F | 6_3 | | 9 | 8325064 | Ames | OU | SetG | G | 7_1 | | 10 | 8325064 | Ames | OU | SetG | G | 7_2 | | 11 | 8325064 | Ames | OU | SetG | G | 7_3 | | 12 | 8325064 | Ames | OU | SetR | R | 18_1 | | 13 | 8325064 | Ames | OU | SetR | R | 18_2 | | 14 | 8325064 | Ames | OU | SetR | R | 18_3 |
|
| Expand |
|---|
| title | gt.xpt (similar to LB) |
|---|
|
| Dataset wrap |
|---|
| | Rowcaps |
|---|
| | Rows 1-7: | Show values collected for the set, SetA. | | Rows 8-15: | Shows values collected for the set, SetF. | | Rows 6-7: | Show a suggested use of the LBSCAT variable. It could be used to further classify types of tests within a laboratory panel (e.g., "DIFFERENTIAL"). | | Rows 6,7,12,13, x,x,x,x: | Show values collected (e.g., MEAN, STANDARD DEVIATION, and/or FOLD INCREASE) that apply to all of the observational units within the experimental unit, as show by OBUID = "ALL". | | Row 9: | Shows the proper use of the LBSTAT variable to indicate "NOT DONE," when the fact that the test was done was documented. | | Row 12: | The subject had Cholesterol measured. The normal range for this test is <200 mg/dL. The value <200 may not be used in the normal range variables LBORNRHI or LBSTNRHI; however, a sponsor may decide, e.g., to enter "0" into LBORNRLO and "199" in LBORNRHI. The sponsor must define the appropriate value for all of the normal range variables. | | Row 13: | Shows the use of LBUSCHFL to indicate that the test result was obtained from an unscheduled blood collection. In this case, the subject was moribund and a blood sample was taken prior to sacrifice. | | Rows 1, 6: | Use records in the SUPPLB dataset example to show biological significance assigned by the investigator for test results. | | Expand |
|---|
| title | Raw Data Table for Study 8325064 |
|---|
|  Image Removed Image Removed
|
| Dataset2 |
|---|
| Row | STUDYID | ASSAYID | DOMAIN | TXSETCD | EUID | OUID | GTSEQ | GTTESTCD | GTTEST | GTORRES | GTORRESU | GTCOLSRT (coll. summ. result type) | GTSTRESC | GTSTRESN | GTSTRESU | GTDTC | GTMETHOD |
|---|
1 | 8325064 | Ames | GT | SETA | A | 0_1 | 1 | RPP | Revertant Numbers Per Plate | 26 | COLONIES | 26 | 26 | COLONIES | 2015-08-03INSTRUMENT COUNTED | 2 | 8325064 | Ames | GT | SETA | A | 0_2 | 2 | RPP | Revertant Numbers Per Plate | 35 | COLONIES | 35 | 35 | 2015-08-03INSTRUMENT COUNTED | 3 | 8325064 | Ames | GT | SETA | A | 0_3 | 3 | RPP | Revertant Numbers Per Plate | 39 | COLONIES | 39 | 39 | 2015-08-03INSTRUMENT COUNTED | 4 | 8325064 | Ames | GT | SETA | A | 0_4 | 4 | RPP | Revertant Numbers Per Plate | 35 | COLONIES | 35 | 35 | 2015-08-03INSTRUMENT COUNTED | 5 | 8325064 | Ames | GT | SETA | A | 0_5 | 5 | RPP | Revertant Numbers Per Plate | 30 | COLONIES | 30 | 30 | 2015-08-03 | INSTRUMENT COUNTED | 6 | 8325064 | Ames | GT | SETA | A | ALL | 6 | RPP | Revertant Numbers Per Plate | 33.0 | COLONIES | MEAN | 33.0 | 33.0 | 7 | 8325064 | Ames | GT | SETA | A | ALL | 7 | RPP | Revertant Numbers Per Plate | 5.0 | COLONIES | STANDARD DEVIATION | 5.0 | 5.0 | 8 | 8325064 | Ames | GT | SETF | F | 6_1 | 1 | RPP | Revertant Numbers Per Plate | 8 | COLONIES | 8 | 8 | MANUALLY COUNTED | 9 | 8325064 | Ames | GT | SETF | F | 6_1 | 2 | CYTOTOX | Cytotoxicity | Very thin background bacterial lawn | COLONIES | V | 10 | 8325064 | Ames | GT | SETF | F | 6_2 | 3 | RPP | Revertant Numbers Per Plate | 10 | COLONIES | 10 | 10 | MANUALLY COUNTED | 11 | 8325064 | Ames | GT | SETF | F | 6_3 | 4 | RPP | Revertant Numbers Per Plate | 12 | COLONIES | 12 | 12 | MANUALLY COUNTED | 12 | 8325064 | Ames | GT | SETF | F | ALL | 5 | RPPMEAN | Mean of Revertant Numbers Per Plate | 10.0 COLONIES | MEAN | 10.0 10.0 13 | 8325064 | Ames | GT | SETF | F | ALL | 6 | RPPSTDDV | Standard Deviation of Revertant Numbers Per Plate | 2.0 | COLONIES | STANDARD DEVIATION | 2.0 | 2.0 | 14 | 8325064 | Ames | GT | SETF | F | ALL | 7 | RPPFDINC | Fold Increase of Revertant Numbers Per Plate | 0.3 | COLONIES | FOLD INCREASE | 0.3 | 0.3 | 8325064 | Ames | GT | SETG | G | 5_1 | 1 | RPP | Revertant Numbers Per Plate | 1104 | 1104 | 1104 | 8325064 | Ames | GT | SETG | G | 5_2 | 2 | RPP | Revertant Numbers Per Plate | 1037 | 1037 | 1037 | 8325064 | Ames | GT | SETG | G | 5_3 | 3 | RPP | Revertant Numbers Per Plate | 971 | 971 | 971 | 8325064 | Ames | GT | SETR | R | ALL | RPPMEAN | Mean of Revertant Numbers Per Plate | 1037.3 1037.3 1037.3 8325064 | Ames | GT | SETR | R | ALL | RPPSTDDV | Standard Deviation of Revertant Numbers Per Plate | 66.5 | 66.5 | 66.5 | 8325064 | Ames | GT | SETR | R | ALL | RPPFLDINC | Fold Increase of Revertant Numbers Per Plate | 31.4 | 31.4 | 31.4
|


