This is an example showing the report table, trial design, and results from a study's in vitro bacterial reverse mutation test
| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-505 |
|---|
|
using 4 different amino acid-requiring strains of
S. typhimurium and 1 strain of
E. coli to detect point mutations.
| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-129 |
|---|
|
In this example, the Trial Summary (TS) dataset| Expand |
|---|
| title | Report Table for Example Study 8325064 |
|---|
|
This table shows the assay results for study 8325064, test article TA1, and 3 strains of salmonella (TA98, TA100, and TA1535) at varying concentrations. For ease of reference, each row has been labeled in the right-hand margin with the applicant-defined trial set label (e.g., SetA). For brevity, the remaining tables (e.g., additional strains, samples prepared without metabolic activation) are not included. | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA- |
|---|
|
|
...
...
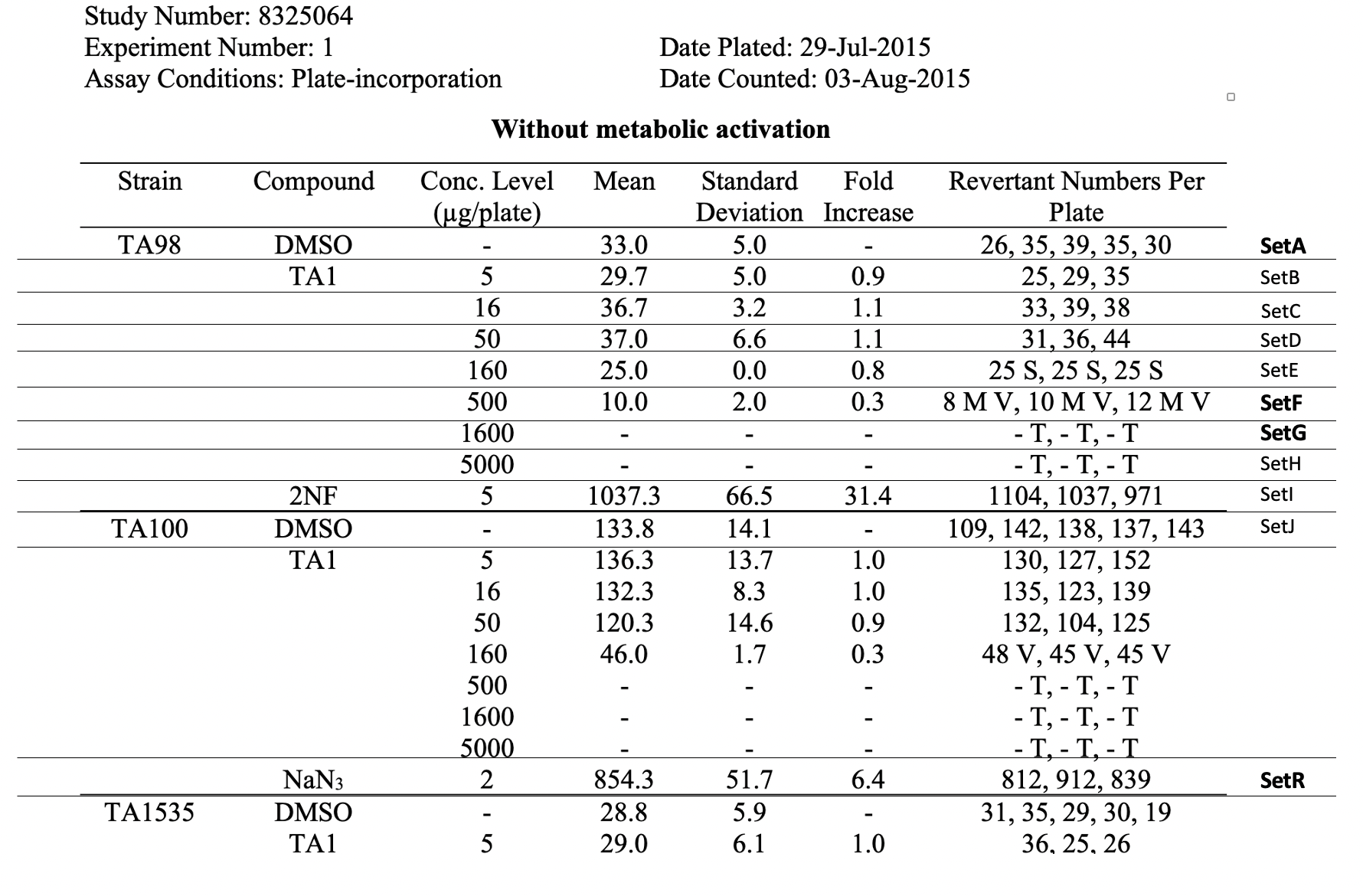 Image Added Image Added
 Image Added Image Added
|
This example Trial Summary (TS) dataset
| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-368 |
|---|
|
includes many informational fields| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-369 |
|---|
|
that may provide context at the study level. ,| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-663 |
|---|
|
shows many informational fields| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-664 |
|---|
|
...
| title | Report Table for Example Study 8325064 |
|---|
that provide context at the study level. ...
| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-663 |
|---|
|
| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA- |
|---|
|
...
 Image Removed
Image Removed
...
| Dataset wrap |
|---|
| Rowcaps |
|---|
| Rows 1-2: | Show 2 records for TSPARMCD = "GLPTYP", using TSSEQ to indicate multiple records, | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-366 |
|---|
|
since both GLP types apply for this study. | | Row 8: | Shows that the applicant's study reference ID is not applicable. | | Rows 9-12: | Show that TSGRPID has been used to link records (name, location, country) related to the test facility (TSGRPID=1). The study director is associated with the test facility. | | Rows 14-16: | Show that TSGRPID (TSGRPID=4) has been used to link the information on the testing guideline followed on this study (TSTGDNAM, TSTGDORG, TSTGDVER). | Jira |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-56 |
|---|
|
| Rows 19, 25: | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-367 |
|---|
|
| Show that this study includes 2 different species of bacteria. | | Rows 18-23: | Show that TSGRPID (TSGRPID = 2) has been used to link the information for 1 species (salmonella) with 4 different strains and the cell line that are tested in this study. | | Rows 25-26: | Show that TSGRPID (TSGRPID = 3) has been used to link the information for 1 species (E. coli) with the strain and cell line that is tested in this study. | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-665 |
|---|
|
|
|
| Dataset2 |
|---|
| Row | STUDYID | GNTXAID | DOMAIN | TSSEQ | TSGRPID | TSPARMCD | TSPARM | TSVAL | TSVALNF |
|---|
| 1 | 8325064 | Ames | TS | 1 |
| GLPTYP | Good Laboratory Practice Type | FDA |
| | 2 | 8325064 | Ames | TS | 2 |
| GLPTYP | Good Laboratory Practice Type | OECD |
| | 3 | 8325064 | Ames | TS | 1 |
| STSTDTC | Study Start Date | 2015-07-29 |
| | 4 | 8325064 | Ames | TS | 1 |
| STITLE | Study Title | The Bacterial Reverse Mutation Test, Study 8325064-1 |
| | 5 | 8325064 | Ames | TS | 1 |
| SNDIGVER | SEND Implementation Guide Version | TOBACCO IMPLEMENTATION GUIDE VERSION 1.0 |
| | 6 | 8325064 | Ames | TS | 1 |
| SNDCTVER | SEND Controlled Terminology Version | SEND Terminology 2021-09-30 |
| | 7 | 8325064 | Ames | TS | 1 |
| SAPPLCNT | Applicant Organization | Example Applicant Inc. |
| | 8 | 8325064 | Ames | TS | 1 |
| SPREFID | Applicant's Study Reference ID |
| NOT APPLICABLE | | 9 | 8325064 | Ames | TS | 1 | 1 | TSTFNAM | Test Facility Name | Example Tox Lab Name |
| | 10 | 8325064 | Ames | TS | 1 | 1 | TSTFLOC | Test Facility Location | 10 Somewhere Street, Montgomery, AL 10000 |
| | 11 | 8325064 | Ames | TS | 1 | 1 | TFCNTRY | Test Facility Country | USA |
| | 12 | 8325064 | Ames | TS | 1 | 1 | STDIR | Study Director | Dr. R. Smith |
| | 13 | 8325064 | Ames | TS | 1 |
| GLPFL | GLP Flag | N |
| | 14 | 8325064 | Ames | TS | 1 | 4 | TSTGDNAM | Testing Guideline Name | Test NO. 471 |
| | 15 | 8325064 | Ames | TS | 1 | 4 | TSTGDORG | Testing Guideline Organization | OECD |
| | 16 | 8325064 | Ames | TS | 1 | 4 | TSTGDVER | Testing Guideline Version | 2020-06-29 |
| | 17 | 8325064 | Ames | TS | 1 |
| SSTYP | Study Type | GENOTOXICITY IN VITRO |
| | 18 | 8325064 | Ames | TS | 1 |
| SSSTYP | Study Sub Type | BACTERIAL REVERSE MUTATION TEST |
| | 19 | 8325064 | Ames | TS | 1 | 2 | SPECIES | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-484 |
|---|
|
| Species | SALMONELLA TYPHIMURIUM | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-123 |
|---|
|
|
| | 20 | 8325064 | Ames | TS | 1 | 2 | STRAIN | Strain/Substrain | TA98 |
| | 21 | 8325064 | Ames | TS | 1 | 2 | STRAIN | Strain/Substrain | TA100 |
| | 22 | 8325064 | Ames | TS | 1 | 2 | STRAIN | Strain/Substrain | TA1535 |
| | 23 | 8325064 | Ames | TS | 1 | 2 | STRAIN | Strain/Substrain | TA1537 |
| | 24 | 8325064 | Ames | TS | 1 | 2 | CELLLN | Cell Line |
|
| | 25 | 8325064 | Ames | TS | 1 | 3 | SPECIES | Species | Escherichia coli | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-482 |
|---|
|
|
| | 26 | 8325064 | Ames | TS | 1 | 3 | STRAIN | Strain/Substrain | WP2 uvrA pKM101 |
| | 27 | 8325064 | Ames | TS | 1 | 3 | CELLLN | Cell Line |
|
|
|
|
...

