This example shows a sample report table, trial design, and results dataset for study 123 for the determination of the in vitro genotoxicity potential of tobacco products using the in vitro micronucleus assay| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-332 |
|---|
|
. | Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
| key | TOBA-135 |
|---|
|
| Jira |
|---|
| showSummary | false |
|---|
| server | Issue Tracker (JIRA) |
|---|
| serverId | 85506ce4-3cb3-3d91-85ee-f633aaaf4a45 |
|---|
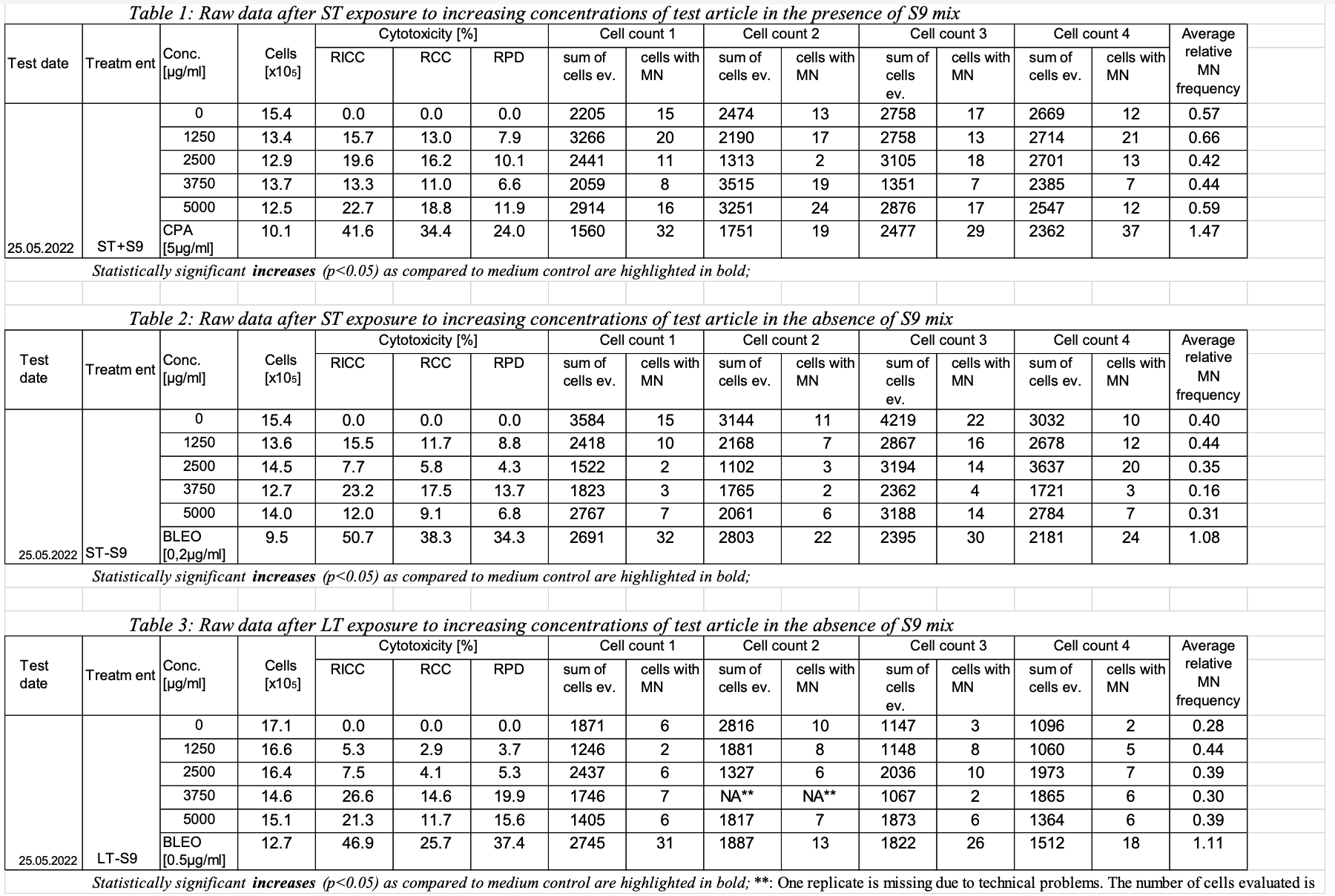
key | TOBA-127| Expand |
|---|
| title | Sample Report Table for Study 123 |
|---|
|
|
...
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
| Rows 1-17: | Show the experimental parameters that are common for table 1 of study 123. | | Rows 18, 25: | Show that sets A1 and A2 each have a parent set code of A. As a child of set A, each of these sets inherit the test conditions of set A. | | Rows 18-24: | Show trial set parameters and values that comprise the test conditions for trial set A1. Set A1 is the data for the negative control (concentration 0) with short-term exposure and metabolic activation S9. The applicant has chosen to given a long name (SET) equal to "ST+S9_C0". Set A1 is associated with the first row in the sample report table for study 123. | | Rows 25-31: | Show trial set parameters and values that comprise the test conditions for trial set A2. Set A2 is the data for the short-term exposure with metabolic activation S9 at a concentration of 1250 ug/ml. The applicant has chosen to give the set a long name (SET) equal to "ST+S9_C1250". Set A2 is associated with the second row in the sample report table for study 123. |
|
| Dataset2 |
|---|
| Row | STUDYID | GNTXAID | DOMAIN | SETCD | SET | TXSEQ | TXPARMCD | TXPARM | TXVAL |
|---|
| 123 | MNvit | TX | A | ST+S9 | | METACT | Metabolic Activation | +S9 | | 2 | 123 | MNvit | TX | A | ST+S9 | 2 | METACTFL | Y/N presence of metabolic activation | Y | | 3 | 123 | MNvit | TX | A | ST+S9 | 3 | TRTDMIN | In vitro Treatment Duration Minimum | 3 | | 4 | 123 | MNvit | TX | A | ST+S9 | 4 | TRTDTRG | In vitro Treatment Duration Target | 3.5 | | 5 | 123 | MNvit | TX | A | ST+S9 | 5 | TRTDMAX | In vitro Treatment Duration Maximum | 4 | | 6 | 123 | MNvit | TX | A | ST+S9 | 6 | TRTDU | In vitro Treatment Duration Unit | HOURS | | 7 | 123 | MNvit | TX | A | ST+S9 | 7 | RCVDMIN | Recovery Duration Minimum | 23.5 | | 8 | 123 | MNvit | TX | A | ST+S9 | 8 | RCVDTRG | Recovery Duration Target | 24 | | 9 | 123 | MNvit | TX | A | ST+S9 | 9 | RCVDMAX | Recovery Duration Maximum | 24.5 | | 10 | 123 | MNvit | TX | A | ST+S9 | 10 | RCVDU | Recovery Duration Unit | HOURS | | 11 | 123 | MNvit | TX | A | ST+S9 | 11 | INCBTMP | Incubation Temperature | 37 | | 12 | 123 | MNvit | TX | A | ST+S9 | 12 | INCBTMPU | Incubation Temperature Unit | C | | 13 | 123 | MNvit | TX | A | ST+S9 | 13 | HUMID | Atmospheric Relative Humidity Percent | 50 | | 14 | 123 | MNvit | TX | A | ST+S9 | 14 | ATMCO2 | Atmospheric CO2 Percent | 5 | | 15 | 123 | MNvit | TX | A | ST+S9 | 15 | SPTOBID | Applicant-defined tobacco identifier | CIG01a | | 16 | 123 | MNvit | TX | A | ST+S9 | 16 | EXPTYP | | Submerged | | 17 | 123 | MNvit | TX | A | ST+S9 | 17 | SAMTYP | Sample Type | Total Particulate Matter in DMSO | | 18 | 123 | MNvit | TX | A1 | ST+S9_C0 | | PSETCD | Parent Set Code | A | | 19 | 123 | MNvit | TX | A1 | ST+S9_CO | 2 | INTRVN | Name of the Intervention Article | Tobacco ProdA | | 20 | 123 | MNvit | TX | A1 | ST+S9_C0 | 3 | ITVTYPE | type of intervention article | Negative Control | | 21 | 123 | MNvit | TX | A1 | ST+S9_C0 | 4 | ITVCONC | Concentration of intervention article | 0 | | 22 | 123 | MNvit | TX | A1 | ST+S9_C0 | 5 | ITVCONCU | Concentration Unit | ug/ml | | 23 | 123 | MNvit | TX | A1 | ST+S9_C0 | 6 | SPDEVID | Applicant-defined device identifier | PUFFMASTER3K | | 24 | 123 | MNvit | TX | A1 | ST+S9_C0 | 7 | DUREFID | Smoke Regimen | Medium Intensity Regimen | | 25 | 123 | MNvit | TX | A2 | ST+S9_C1250 | | PSETCD | Parent Set Code | A | | 26 | 123 | MNvit | TX | A2 | ST+S9_C1250 | 2 | INTRVN | Name of the Intervention Article | Tobacco ProdA | | 27 | 123 | MNvit | TX | A2 | ST+S9_C1250 | 3 | ITVTYPE | type of intervention article | Product | | 28 | 123 | MNvit | TX | A2 | ST+S9_C1250 | 4 | ITVCONC | Concentration of intervention article | 1250 | | 29 | 123 | MNvit | TX | A2 | ST+S9_C1250 | 5 | ITVCONCU | Concentration Unit | ug/ml | | 30 | 123 | MNvit | TX | A2 | ST+S9_C1250 | 6 | SPDEVID | Applicant-defined device identifier | PUFFMASTER2023 | | 31 | 123 | MNvit | TX | A2 | ST+S9_C1250 | 7 | DUREFID | Smoke Regimen | High Intensity Regimen |
|
|
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
| Row 1: | Shows the value of REFID=C0. This REFID refers to the trial set with a SETCD of "A1", as defined in the TX dataset. LEVEL=1 and LVLDESC="EXPERIMENTAL UNIT/TRIAL SET" indicates this identifier is referring to both the experimental unit and the unit to which the treatment is applied, and to the entire trial set. | | Rows 2-5: | Show the values of 4 observational units (C0_Count1 through C0_Count4) that are within the parent experimental unit, REFID=C0. In this example assay, these observational units are also all within the same trial set, as defined in the TX dataset. | | Row 6: | Shows the value of REFID=C1250. This REFID refers to the trial set with a SETCD of "A2", as defined in the TX dataset. LEVEL=1 and LVLDESC="EXPERIMENTAL UNIT/TRIAL SET" indicates this identifier is referring to both the experimental unit and the unit to which the treatment is applied, and to the entire trial set. | | Rows 7-10: | Show the values of 4 observational units (C1250_Count1 through C1250_Count4) that are within the parent experimental unit, REFID=C0. In this example assay, these observational units are also all within the same trial set, as defined in the TX dataset. |
|
| Dataset2 |
|---|
| Row | STUDYID | GNTXAID | SETCD | REFID | PARENT | LEVEL | LVLDESC |
|---|
1 | 123 | MNvit | A1 | C0 |
| 1 | EXPERIMENTAL UNIT/TRIAL SET | 2 | 123 | MNvit | A1 | C0-Count1 | C0 | 2 | OBSERVATIONAL UNIT | 3 | 123 | MNvit | A1 | C0-Count2 | C0 | 2 | OBSERVATIONAL UNIT | 4 | 123 | MNvit | A1 | C0-Count3 | C0 | 2 | OBSERVATIONAL UNIT | 5 | 123 | MNvit | A1 | C0-Count4 | C0 | 2 | OBSERVATIONAL UNIT | 6 | 123 | MNvit | A2 | C1250 |
| 1 | EXPERIMENTAL UNIT/TRIAL SET | | 7 | 123 | MNvit | A2 | C1250-Count1 | C1250 | 2 | OBSERVATIONAL UNIT | | 8 | 123 | MNvit | A2 | C1250-Count2 | C1250 | 2 | OBSERVATIONAL UNIT | | 9 | 123 | MNvit | A2 | C1250-Count3 | C1250 | 2 | OBSERVATIONAL UNIT | | 10 | 123 | MNvit | A2 | C1250-Count4 | C1250 | 2 | OBSERVATIONAL UNIT |
|
|
| Dataset wrap |
|---|
|
| Rowcaps |
|---|
Rows 1-3, 8: | Show percentage result values that apply to GTREFID=C0. REFID=C0, as shown in the RELREF dataset, relates this data to the trial set in the first row of table 1 in the sample report table for study 123. | | Rows 4-7: | Show the 4 micronucleated cell counts for the observational units with GTREFID from C0-Count1 through C0-Count4, for which their relationship to test conditions (in tx.xpt) and experimental units (in relref.xpt) are shown in the RELREF dataset. | Rows 9-11, 16: | Show percentage result values that apply to GTREFID=C1250. REFID=C1250, as shown in the RELREF dataset, relates this data to the trial set in the second row of table 1 in the sample report table for study 123. | | Rows 12-15: | Show the 4 micronucleated cell counts for the observational units with GTREFID from C1250-Count1 through C1250-Count4, for which their relationship to test conditions (in tx.xpt) and experimental units (in relref.xpt) are shown in the RELREF dataset. |
|
| Dataset2 |
|---|
| Row | STUDYID | GNTXAID | DOMAIN | GTREFID | GTSEQ | GTTESTCD | GTTEST | GTCELLEV | GTORRES | GTORRESU | GTCOLSRT | GTSTRESC | GTSTRESN | GTSTRESU | GTDTC |
|---|
| 1 | 123 | MNvit | GT | C0 | 1 | RICC | Relative Increase in Cell Count | 154 | 0 | % |
| 0 | 0 | % | 2022-05-25 | | 2 | 123 | MNvit | GT | C0 | 2 | RCC | Relative Cell Count | 154 | 0 | % |
| 0 | 0 | % | 2022-05-25 | | 3 | 123 | MNvit | GT | C0 | 3 | RPD | Relative Population Doubling | 154 | 0 | % |
| 0 | 0 | % | 2022-05-25 | | 4 | 123 | MNvit | GT | C0-Count1 | 4 | MNCE | Micronucleated Cells | 2205 | 15 | Cells |
| 15 | 15 | Cells | 2022-05-25 | | 5 | 123 | MNvit | GT | C0-Count2 | 5 | MNCE | Micronucleated Cells | 2474 | 13 | Cells |
| 13 | 13 | Cells | 2022-05-25 | | 6 | 123 | MNvit | GT | C0-Count3 | 6 | MNCE | Micronucleated Cells | 2758 | 17 | Cells |
| 17 | 17 | Cells | 2022-05-25 | | 7 | 123 | MNvit | GT | C0-Count4 | 7 | MNCE | Micronucleated Cells | 2669 | 12 | Cells |
| 12 | 12 | Cells | 2022-05-25 | | 8 | 123 | MNvit | GT | C0 | 8 | MNCECE | Micronucleated Cells/Total Cells |
| 0.57 | % |
| 0.57 | 0.57 | % | 2022-05-25 | | 9 | 123 | MNvit | GT | C1250 | 1 | RICC | Relative Increase in Cell Count | 134 | 15.7 | % |
| 15.7 | 15.7 | % | 2022-05-25 | | 10 | 123 | MNvit | GT | C1250 | 2 | RCC | Relative Cell Count | 134 | 13.0 | % |
| 13.0 | 13.0 | % | 2022-05-25 | | 11 | 123 | MNvit | GT | C1250 | 3 | RPD | Relative Population Doubling | 134 | 7.9 | % |
| 7.9 | 7.9 | % | 2022-05-25 | | 12 | 123 | MNvit | GT | C1250-Count1 | 4 | MNCE | Micronucleated Cells | 3266 | 20 | Cells |
| 20 | 20 | Cells | 2022-05-25 | | 13 | 123 | MNvit | GT | C1250-Count2 | 5 | MNCE | Micronucleated Cells | 2190 | 17 | Cells |
| 17 | 17 | Cells | 2022-05-25 | | 14 | 123 | MNvit | GT | C1250-Count3 | 6 | MNCE | Micronucleated Cells | 2758 | 13 | Cells |
| 13 | 13 | Cells | 2022-05-25 | | 15 | 123 | MNvit | GT | C1250-Count4 | 7 | MNCE | Micronucleated Cells | 2714 | 21 | Cells |
| 21 | 21 | Cells | 2022-05-25 | | 16 | 123 | MNvit | GT | C1250 | 8 | MNCECE | Micronucleated Cells/Total Cells |
| 0.66 | % |
| 0.66 | 0.66 | % | 2022-05-25 |
|
|
