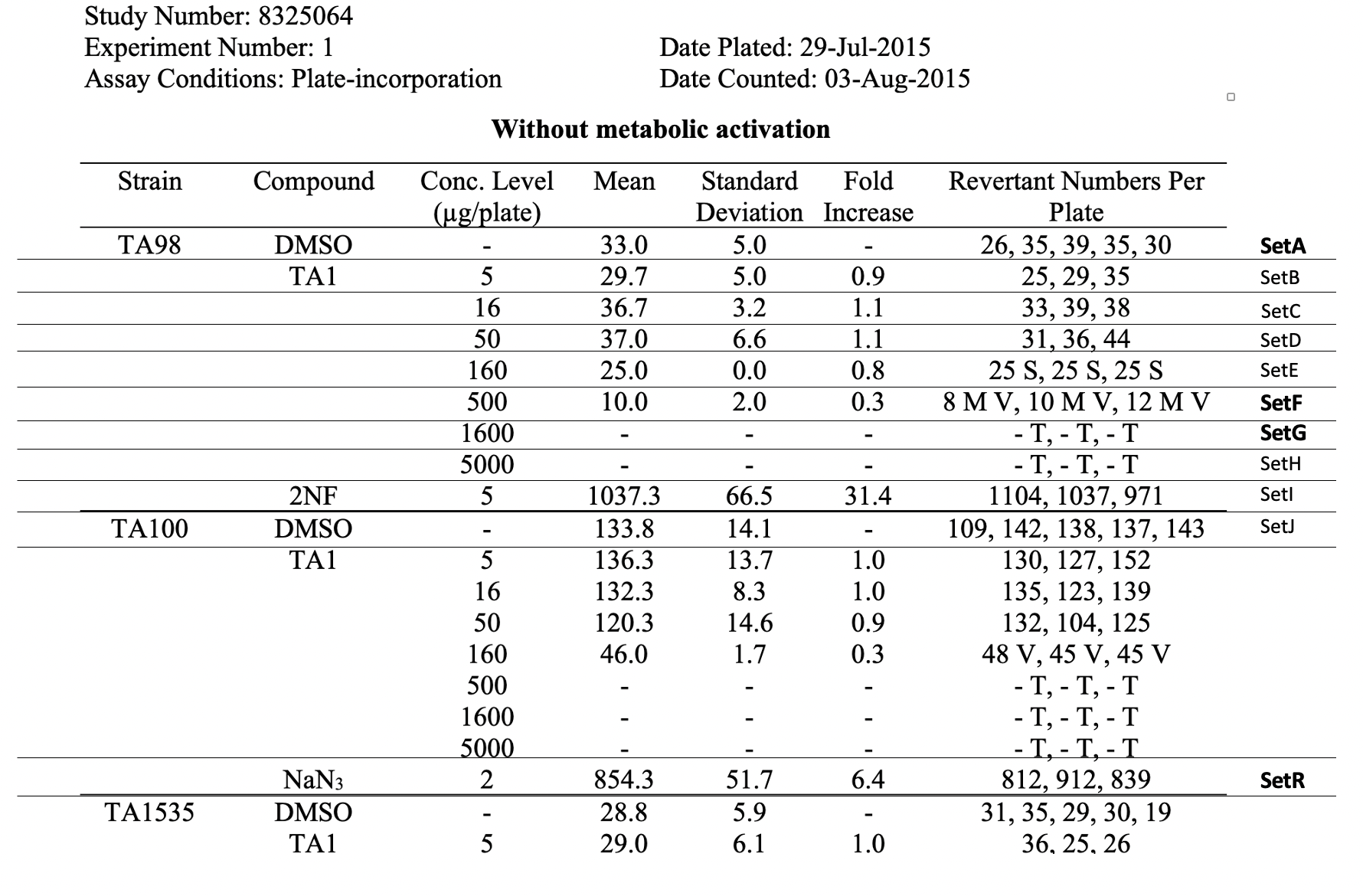
| Dataset wrap |
|---|
| Rowcaps |
|---|
NOTE: Observational Unit Identifiers (OBUIDs) are defined by the sponsor to uniquely identify the observational unit within an experimental unit. In this example OBUIDs are defined by the sponsor based on the test conditions (trial set) and the unit's location on one of multiple exposure/incubation plates. | Rows 1-5: | Show the values for OBUID within the experimental unit identifier for Set A, as defined in the trial set dataset. | | Rows 6-8: | Show the values for OBUID within the experimental unit identifier for Set F, as defined in the trial set dataset |
|
|
Experimental unit - The unit to which the treatment is applied, eg. Test tube Observational unit - The unit on which the response is measured. (This may not be the same as the experimental unit.) | Rowcaps |
|---|
| Dataset wrap |
|---|
| | Rows 1-5: | Shows the values of the experimental unit identifiers and associated observational unit identifiers for the set, SetA.
SetA is the first row of the raw data table, see the row labeled "SetA" in the table, Raw Data for Study 8325064. | Rows 6-8: | Shows the values of the experimental unit identifiers and associated observational unit identifiers for the set, SetF.
SetF is the sixth row of the raw data table, see the row labeled "SetF" in the table, Raw Data for Study 8325064Shows of | for OBUID within the experimental unit |
|
identifiers and associated observational unit identifiers for the set, SetG.
SetG is the seventh row of the raw data table, see the row labeled "SetG" in the table, Raw Data for Study 8325064| identifier for Set G, as defined in the trial set dataset. | | Row 12-14: |
|
Shows of | for OBUID within the experimental unit |
|
identifiers and associated observational unit identifiers for the set, SetR.
SetR is the eighteenth row of the raw data table, see the row labeled "SetR" in the table, Raw Data for Study 8325064.Rows 2-4: | Rows 1, 6: |
Observational Unit Identifiers (OBUIDs) are defined by the sponsor to uniquely identify the observational unit within an experimental unit. In this example OBUIDs are defined by the sponsor based on the test conditions (trial set) and the unit's location on one of multiple exposure/incubation plates. | Rowcaps |
|---|
| Rows 1-5: | Show the values for OBUID within the experimental unit identifier for Set A, as defined in the trial set dataset. | | Rows 6-8: | Show the values for OBUID within the experimental unit identifier for Set F, as defined in the trial set dataset. | | Rows 9-11: | Show the values for OBUID within the experimental unit identifier for Set G, as defined in the trial set dataset. | | Row 12-14: | Show the values for OBUID within the experimental unit identifier for Set R, as defined in the trial set dataset. | | Dataset2 |
|---|
| Row | STUDYID | ASSAYID | DOMAIN | SETCD | EUID | OBUID |
|---|
| 1 | 8325064 | Ames | OU | SetA | A | 0_1 | | 2 | 8325064 | Ames | OU | SetA | A | 0_2 | | 3 | 8325064 | Ames | OU | SetA | A | 0_3 | | 4 | 8325064 | Ames | OU | SetA | A | 0_4 | | 5 | 8325064 | Ames | OU | SetA | A | 0_5 | 6 | 8325064 | Ames | OU | SetF | F | 6_1 | | 7 | 8325064 | Ames | OU | SetF | F | 6_2 | | 8 | 8325064 | Ames | OU | SetF | F | 6_3 | | 9 | 8325064 | Ames | OU | SetG | G | 7_1 | | 10 | 8325064 | Ames | OU | SetG | G | 7_2 | | 11 | 8325064 | Ames | OU | SetG | G | 7_3 | | 12 | 8325064 | Ames | OU | SetR | R | 18_1 | | 13 | 8325064 | Ames | OU | SetR | R | 18_2 | 14 | 8325064 | Ames | OU | SetR | R | 18_3| identifier for Set R, as defined in the trial set dataset. |
|
| Dataset2 |
|---|
| Row | STUDYID | ASSAYID | DOMAIN | SETCD | EUID | OBUID |
|---|
| 1 | 8325064 | Ames | OU | SetA | A | 0_1 | | 2 | 8325064 | Ames | OU | SetA | A | 0_2 | | 3 | 8325064 | Ames | OU | SetA | A | 0_3 | | 4 | 8325064 | Ames | OU | SetA | A | 0_4 | | 5 | 8325064 | Ames | OU | SetA | A | 0_5 | 6 | 8325064 | Ames | OU | SetF | F | 6_1 | | 7 | 8325064 | Ames | OU | SetF | F | 6_2 | | 8 | 8325064 | Ames | OU | SetF | F | 6_3 | | 9 | 8325064 | Ames | OU | SetG | G | 7_1 | | 10 | 8325064 | Ames | OU | SetG | G | 7_2 | | 11 | 8325064 | Ames | OU | SetG | G | 7_3 | | 12 | 8325064 | Ames | OU | SetR | R | 18_1 | | 13 | 8325064 | Ames | OU | SetR | R | 18_2 | | 14 | 8325064 | Ames | OU | SetR | R | 18_3 |
|
|
|